DNA ligands that bind tightly and selectively to cellobiose
- PMID: 9576904
- PMCID: PMC20399
- DOI: 10.1073/pnas.95.10.5462
DNA ligands that bind tightly and selectively to cellobiose
Abstract
Cell surface oligosaccharides have been shown to play essential biological roles in such diverse biological phenomena as cellular adhesion, molecular recognition, and inflammatory response. The development of high-affinity ligands capable of selectively recognizing a variety of small motifs in different oligosaccharides would be of significant interest as experimental and diagnostic tools. As a step toward this goal we have developed DNA ligands that recognize the disaccharide cellobiose, whether in soluble form or as the repeating unit of the polymer, cellulose. These DNA "aptamers" bind with high selectivity to cellobiose with little or no affinity for the related disaccharides lactose, maltose, and gentiobiose. Thus, the DNA ligands can discriminate sugar epimers, anomers, and disaccharide linkages.
Figures
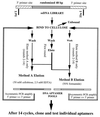
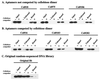
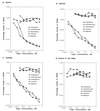

Similar articles
-
Investigation of disaccharide recognition by molecularly imprinted polymers.Bioseparation. 2001;10(6):307-14. doi: 10.1023/a:1021589619501. Bioseparation. 2001. PMID: 12549874
-
Molecular determinants of ligand specificity in family 11 carbohydrate binding modules: an NMR, X-ray crystallography and computational chemistry approach.FEBS J. 2008 May;275(10):2524-35. doi: 10.1111/j.1742-4658.2008.06401.x. Epub 2008 Apr 15. FEBS J. 2008. PMID: 18422658
-
Utilization of intravenously administered beta-cellobiose and maltose by young pigs.J Nutr. 1983 May;113(5):1039-45. doi: 10.1093/jn/113.5.1039. J Nutr. 1983. PMID: 6842298
-
[Regulation of the formation and conversion of intermediate cellooligosaccharides and cellobiose during ezymatic hydrolysis of insoluble cellulose].Biokhimiia. 1982 Apr;47(4):608-18. Biokhimiia. 1982. PMID: 7200805 Russian.
-
A novel transcriptional regulator, ClbR, controls the cellobiose- and cellulose-responsive induction of cellulase and xylanase genes regulated by two distinct signaling pathways in Aspergillus aculeatus.Appl Microbiol Biotechnol. 2013 Mar;97(5):2017-28. doi: 10.1007/s00253-012-4305-8. Epub 2012 Aug 1. Appl Microbiol Biotechnol. 2013. PMID: 22851016
Cited by
-
Aptamers: versatile molecular recognition probes for cancer detection.Analyst. 2016 Jan 21;141(2):403-15. doi: 10.1039/c5an01995h. Analyst. 2016. PMID: 26618445 Free PMC article. Review.
-
Functional nucleic acid sensors.Chem Rev. 2009 May;109(5):1948-98. doi: 10.1021/cr030183i. Chem Rev. 2009. PMID: 19301873 Free PMC article. Review. No abstract available.
-
Targeting Oligosaccharides and Glycoconjugates Using Superselective Binding Scaffolds.Adv Funct Mater. 2020 Aug 3;30(31):2002298. doi: 10.1002/adfm.202002298. Epub 2020 May 28. Adv Funct Mater. 2020. PMID: 32774200 Free PMC article.
-
Single-stranded DNA oligoaptamers: molecular recognition and LPS antagonism are length- and secondary structure-dependent.J Innate Immun. 2009;1(1):46-58. doi: 10.1159/000145542. Epub 2008 Jul 12. J Innate Immun. 2009. PMID: 20375565 Free PMC article.
-
Acyclic identification of aptamers for human alpha-thrombin using over-represented libraries and deep sequencing.PLoS One. 2011;6(5):e19395. doi: 10.1371/journal.pone.0019395. Epub 2011 May 19. PLoS One. 2011. PMID: 21625587 Free PMC article.
References
-
- Goldstein I J, Poretz R D. In: The Lectins: Properties, Function, and Applications in Biology and Medicine. Liener I E, Sharon N, Goldstein I J, editors. San Diego: Academic; 1986. pp. 33–247.
-
- Weis W I, Drickamer K. Annu Rev Biochem. 1996;65:441–473. - PubMed
-
- Gabius H-J. Eur J Biochem. 1997;243:543–576. - PubMed
-
- Glaudemans C P J. Chem Rev. 1991;91:25–33.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

