The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors
- PMID: 9576906
- PMCID: PMC20401
- DOI: 10.1073/pnas.95.10.5474
The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors
Abstract
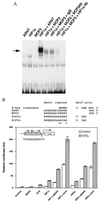
We report that MOP3 is a general dimerization partner for a subset of the basic-helix-loop-helix (bHLH)-PER-ARNT-SIM (PAS) superfamily of transcriptional regulators. We demonstrated that MOP3 interacts with MOP4, CLOCK, hypoxia-inducible factor 1alpha (HIF1alpha), and HIF2alpha. A DNA selection protocol revealed that the MOP3-MOP4 heterodimer bound a CACGTGA-containing DNA element. Transient transfection experiments demonstrated that the MOP3-MOP4 and MOP3-CLOCK complexes bound this element in COS-1 cells and drove transcription from a linked luciferase reporter gene. We also deduced the high-affinity DNA binding sites for MOP3-HIF1alpha complex (TACGTGA) and used transient transfection experiments to demonstrate that the MOP3-HIF1alpha and MOP3-HIF2alpha heterodimers bound this element, drove transcription, and responded to cellular hypoxia. Finally, we found that MOP3 mRNA expression overlaps in a number of tissues with each of its four potential partner molecules in vivo.
Figures



References
-
- Huang Z J, Edery I, Rosbash M. Nature (London) 1993;364:259–262. - PubMed
-
- Hoffman E C, Reyes H, Chu F F, Sander F, Conley L H, Brooks B A, Hankinson O. Science. 1991;252:954–958. - PubMed
-
- Nambu J R, Lewis J O, Wharton K A, Jr, Crews S T. Cell. 1991;67:1157–1167. - PubMed
-
- Jackson F R, Bargiello T A, Yun S H, Young M W. Nature (London) 1986;320:185–188. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

