MMS2, encoding a ubiquitin-conjugating-enzyme-like protein, is a member of the yeast error-free postreplication repair pathway
- PMID: 9576943
- PMCID: PMC20438
- DOI: 10.1073/pnas.95.10.5678
MMS2, encoding a ubiquitin-conjugating-enzyme-like protein, is a member of the yeast error-free postreplication repair pathway
Abstract
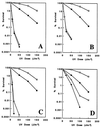
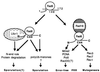
Among the three Saccharomyces cerevisiae DNA repair epistasis groups, the RAD6 group is the most complicated and least characterized, primarily because it consists of two separate repair pathways: an error-free postreplication repair pathway, and a mutagenesis pathway. The rad6 and rad18 mutants are defective in both pathways, and the rev3 mutant affects only the mutagenesis pathway, but a yeast gene that is involved only in error-free postreplication repair has not been reported. We cloned the MMS2 gene from a yeast genomic library by functional complementation of the mms2-1 mutant [Prakash, L. & Prakash, S. (1977) Genetics 86, 33-55]. MMS2 encodes a 137-amino acid, 15.2-kDa protein with significant sequence homology to a conserved family of ubiquitin-conjugating (Ubc) proteins. However, Mms2 does not appear to possess Ubc activity. Genetic analyses indicate that the mms2 mutation is hypostatic to rad6 and rad18 but is synergistic with the rev3 mutation, and the mms2 mutant is proficient in UV-induced mutagenesis. These phenotypes are reminiscent of a pol30-46 mutant known to be impaired in postreplication repair. The mms2 mutant also displayed a REV3-dependent mutator phenotype, strongly suggesting that the MMS2 gene functions in the error-free postreplication repair pathway, parallel to the REV3 mutagenesis pathway. Furthermore, with respect to UV sensitivity, mms2 was found to be hypostatic to the rad6Delta1-9 mutation, which results in the absence of the first nine amino acids of Rad6. On the basis of these collective results, we propose that the mms2 null mutation and two other allele-specific mutations, rad6Delta1-9 and pol30-46, define the error-free mode of DNA postreplication repair, and that these mutations may enhance both spontaneous and DNA damage-induced mutagenesis.
Figures



References
-
- Finley D, Bartel B, Varshavsky A. Nature (London) 1989;338:394–401. - PubMed
-
- Chen P, Johnson P, Sommer T, Jentsch S, Hochstrasser M. Cell. 1993;74:357–369. - PubMed
-
- Gobel M G, Yochem J, Jentsch S, McGrath J P, Varshavsky A, Byers B. Science. 1988;241:1331–1335. - PubMed
-
- Jentsch S, McGrath J P, Varshavsky A. Nature (London) 1987;329:131–134. - PubMed
-
- Finley D, Ozkaynak E, Varshavsky A. Cell. 1987;48:1035–1046. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

