Alternative splicing of rearranged T cell receptor delta sequences to the constant region of the alpha locus
- PMID: 9576946
- PMCID: PMC20441
- DOI: 10.1073/pnas.95.10.5694
Alternative splicing of rearranged T cell receptor delta sequences to the constant region of the alpha locus
Abstract
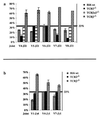
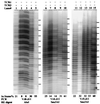
The T cell receptor (TCR) alpha/delta locus is composed of a common, shared set of variable (V) and distinct diversity (D), joining (J), and constant (C) genes. It has been recognized for several years that transcripts of the rearranged VDJdelta or VJalpha genes are spliced to the Cdelta or Calpha genes, respectively, encoding distinct TCR delta and alpha proteins. Herein, we describe the discovery of a splicing variation that allows the assembled VDJdelta genes to be fused with the Calpha gene. This variation is prominent in TCRdelta gene-deficient mice but is also detectable in wild-type mice. Furthermore, we show that several in-frame VDJdelta rearrangements in TCRdelta gene-deficient mice are strikingly underrepresented, suggesting that the alternative transcripts, with protein coding capacity, influence the development of alphabeta thymocytes. In-frame TCRgamma gene rearrangements do not appear underrepresented, indicating that the effect is not mediated by the gamma chain. Instead, indirect evidence supports the hypothesis that the delta/alpha chimeric protein acts in conjunction with the TCRbeta chain. These results have implications for the transcriptional control of the TCRalpha/delta locus and provide a novel insight into the distinct functional capacities of the TCR alpha and delta proteins during thymocyte development.
Figures


References
-
- Schatz D G, Oettinger M A, Schlissel M S. Annu Rev Immunol. 1992;10:359–383. - PubMed
-
- Chien Y, Iwashima M, Kaplan K B, Elliot J F, Davis M M. Nature (London) 1987;327:677–682. - PubMed
-
- Wang K, Klotz J L, Kiser G, Bristol G, Hays E, Lai E, Gese E, Kronenberg M, Hood L. Genomics. 1994;20:419–428. - PubMed
-
- Kronenberg M, Siu G, Hood L, Shastri N. Annu Rev Immunol. 1986;4:529–591. - PubMed
-
- Koop B F, Wilson R K, Wang K, Vernooij B, Zallwer D, Kuo C, Seto D, Toda M, Hood L. Genomics. 1992;13:1209–1230. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

