Isolation of a hepadnavirus from the woolly monkey, a New World primate
- PMID: 9576957
- PMCID: PMC20452
- DOI: 10.1073/pnas.95.10.5757
Isolation of a hepadnavirus from the woolly monkey, a New World primate
Abstract
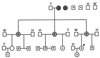
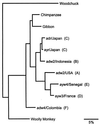
Hepatitis B virus (HBV) infections are a major worldwide health problem with chronic infections leading to cirrhosis and liver cancer. Viruses related to human HBV have been isolated from birds and rodents, but despite efforts to find hepadnaviruses that infect species intermediate in evolution between rodents and humans, none have been described. We recently isolated a hepadnavirus from a woolly monkey (Lagothrix lagotricha) that was suffering from fulminant hepatitis. Phylogenetic analysis of the nucleotide sequences of the core and surface genes indicated that the virus was distinct from the human HBV family, and because it is basal (ancestral) to the human monophyletic group, it probably represents a progenitor of the human viruses. This virus was designated woolly monkey hepatitis B virus (WMHBV). Analysis of woolly monkey colonies at five zoos indicated that WMHBV infections occurred in most of the animals at the Louisville zoo but not at four other zoos in the United States. The host range of WMHBV was examined by inoculation of one chimpanzee and two black-handed spider monkeys (Ateles geoffroyi), the closest nonendangered relative of the woolly monkey. The data suggest that spider monkeys are susceptible to infection with WMHBV and that minimal replication was observed in a chimpanzee. Thus, we have isolated a hepadnavirus with a host intermediate between humans and rodents and establishes a new animal model for evaluation of antiviral therapies for treating HBV chronic infections.
Figures
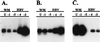



References
-
- Beasley R P, Hwang L-Y, Lin C-C, Chien C-S. Lancet. 1981;ii:1129–1133. - PubMed
-
- Young P. ASM News. 1996;62:450–451.
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

