GFRalpha3 is an orphan member of the GDNF/neurturin/persephin receptor family
- PMID: 9576965
- PMCID: PMC20460
- DOI: 10.1073/pnas.95.10.5801
GFRalpha3 is an orphan member of the GDNF/neurturin/persephin receptor family
Abstract
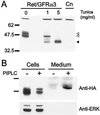
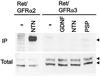
GDNF, neurturin, and persephin are transforming growth factor beta-related neurotrophic factors known collectively as the GDNF family (GF). GDNF and neurturin signal through a multicomponent receptor complex containing a signaling component (the Ret receptor tyrosine kinase) and either of two glycosyl-phosphatidylinositol-linked binding components (GDNF family receptor alpha components 1 and 2, GFRalpha1 or GFRalpha2), whereas the receptor for persephin is unknown. Herein we describe a third member of the GF coreceptor family called GFRalpha3 that is encoded by a gene located on human chromosome 5q31.2-32. GFRalpha3 is not expressed in the central nervous system of the developing or adult animal but is highly expressed in several developing and adult sensory and sympathetic ganglia of the peripheral nervous system. GFRalpha3 is also expressed at high levels in developing, but not adult, peripheral nerve. GFRalpha3 is a glycoprotein that is glycosyl-phosphatidylinositol-linked to the cell surface like GFRalpha1 and GFRalpha2. Fibroblasts expressing Ret and GFRalpha3 do not respond to any of the known members of the GDNF family, suggesting that GFRalpha3 interacts with an unknown ligand or requires a different or additional signaling protein to function.
Figures



References
-
- Lin L-F H, Doherty D H, Lile J D, Bektesh S, Collins F. Science. 1993;260:1130–1132. - PubMed
-
- Kotzbauer P T, Lampe P A, Heuckeroth R O, Golden J P, Creedon D J, Johnson E M, Milbrandt J D. Nature (London) 1996;384:467–470. - PubMed
-
- Milbrandt J, de Sauvage F, Fahrner T J, Baloh R H, Leitner M L, Tansey M G, Lampe P A, Heuckeroth R O, Kotzbauer P T, Simburger K S, et al. Neuron. 1998;20:245–253. - PubMed
-
- Stromberg I, Bjorklund L, Johansson M, Tomac A, Collins F, Olson L, Hoffer B, Humpel C. Exp Neurol. 1993;124:401–412. - PubMed
-
- Hudson J, Granholm A-C, Gerhardt G A, Henry M A, Hoffman A, Biddle P, Leela N S, Mackerlova L, Lile J D, Collins F, Hoffer B J. Brain Res Bull. 1995;36:425–432. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

