The regulatory particle of the Saccharomyces cerevisiae proteasome
- PMID: 9584156
- PMCID: PMC108897
- DOI: 10.1128/MCB.18.6.3149
The regulatory particle of the Saccharomyces cerevisiae proteasome
Abstract
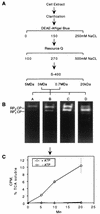
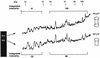
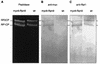
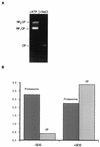
The proteasome is a multisubunit protease responsible for degrading proteins conjugated to ubiquitin. The 670-kDa core particle of the proteasome contains the proteolytic active sites, which face an interior chamber within the particle and are thus protected from the cytoplasm. The entry of substrates into this chamber is thought to be governed by the regulatory particle of the proteasome, which covers the presumed channels leading into the interior of the core particle. We have resolved native yeast proteasomes into two electrophoretic variants and have shown that these represent core particles capped with one or two regulatory particles. To determine the subunit composition of the regulatory particle, yeast proteasomes were purified and analyzed by gradient sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Resolution of the individual polypeptides revealed 17 distinct proteins, whose identities were determined by amino acid sequence analysis. Six of the subunits have sequence features of ATPases (Rpt1 to Rpt6). Affinity chromatography was used to purify regulatory particles from various strains, each of which expressed one of the ATPases tagged with hexahistidine. In all cases, multiple untagged ATPases copurified, indicating that the ATPases assembled together into a heteromeric complex. Of the remaining 11 subunits that we have identified (Rpn1 to Rpn3 and Rpn5 to Rpn12), 8 are encoded by previously described genes and 3 are encoded by genes not previously characterized for yeasts. One of the previously unidentified subunits exhibits limited sequence similarity with deubiquitinating enzymes. Overall, regulatory particles from yeasts and mammals are remarkably similar, suggesting that the specific mechanistic features of the proteasome have been closely conserved over the course of evolution.
Figures






References
-
- Adams G M, Falke S, Goldberg A L, Slaughter C A, DeMartino G N, Gogol E P. Structural and functional effects of PA700 and modulator protein on proteasomes. J Mol Biol. 1997;273:646–657. - PubMed
-
- Akiyama K, Yokota K, Kagawa S, Shimbara N, DeMartino G N, Slaughter C A, Noda C, Tanaka K. cDNA cloning of a new putative ATPase subunit p45 of the human 26S proteasome, a homolog of yeast Sug1. FEBS Lett. 1995;363:151–156. - PubMed
-
- Armon T, Ganoth D, Hershko A. Assembly of the 26S complex that degrades proteins ligated to ubiquitin is accompanied by the formation of ATPase activity. J Biol Chem. 1990;265:20723–20726. - PubMed
-
- Baker R T, Tobias J W, Varshavsky A. Ubiquitin-specific proteases of S. cerevisiae. Cloning of UBP2 and UBP3 and functional analysis of the UBPgene family. J Biol Chem. 1992;267:23364–23375. - PubMed
-
- Bauer V W, Swaffield J C, Johnston S A, Andrews M T. CADp44: a novel regulatory subunit of the 26S proteasome and the mammalian homolog of ySug2p. Gene. 1996;181:63–69. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
