XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage
- PMID: 9584196
- PMCID: PMC108937
- DOI: 10.1128/MCB.18.6.3563
XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage
Abstract
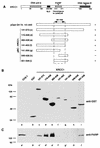
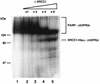
Poly(ADP-ribose) polymerase (PARP; EC 2.4.2.30) is a zinc-finger DNA-binding protein that detects and signals DNA strand breaks generated directly or indirectly by genotoxic agents. In response to these breaks, the immediate poly(ADP-ribosyl)ation of nuclear proteins involved in chromatin architecture and DNA metabolism converts DNA damage into intracellular signals that can activate DNA repair programs or cell death options. To have greater insight into the physiological function of this enzyme, we have used the two-hybrid system to find genes encoding proteins putatively interacting with PARP. We have identified a physical association between PARP and the base excision repair (BER) protein XRCC1 (X-ray repair cross-complementing 1) in the Saccharomyces cerevisiae system, which was further confirmed to exist in mammalian cells. XRCC1 interacts with PARP by its central region (amino acids 301 to 402), which contains a BRCT (BRCA1 C terminus) module, a widespread motif in DNA repair and DNA damage-responsive cell cycle checkpoint proteins. Overexpression of XRCC1 in Cos-7 or HeLa cells dramatically decreases PARP activity in vivo, reinforcing the potential protective function of PARP at DNA breaks. Given that XRCC1 is also associated with DNA ligase III via a second BRCT module and with DNA polymerase beta, our results provide strong evidence that PARP is a member of a BER multiprotein complex involved in the detection of DNA interruptions and possibly in the recruitment of XRCC1 and its partners for efficient processing of these breaks in a coordinated manner. The modular organizations of these interactors, associated with small conserved domains, may contribute to increasing the efficiency of the overall pathway.
Figures





References
-
- Althaus F R, Richter C. ADP-ribosylation of proteins. Enzymology and biological significance. Mol Biol Biochem Biophys. 1987;37:1–237. - PubMed
-
- Barlow C, Hirotsune S, Paylor R, Liyanage M, Eckhaus M, Collins F, Shilo Y, Crawley J N, Ried T, Tagle D, Wynshaw-Boris A. Atm-deficient mice: a paradigm of ataxia telangectasia. Cell. 1996;86:159–171. - PubMed
-
- Bork P, Hofmann K, Bucher P, Neuwald A F, Altschul S F, Koonin E V. A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J. 1997;11:68–76. - PubMed
-
- Brookman K W, Tebbs R S, Allen S A, Tucker J D, Swiger R R, Lamerdin J E, Carrano A V, Thompson L H. Isolation and characterization of mouse Xrcc-1, a DNA repair gene affecting ligation. Genomics. 1994;22:180–188. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
