Spalten, a protein containing Galpha-protein-like and PP2C domains, is essential for cell-type differentiation in Dictyostelium
- PMID: 9585512
- PMCID: PMC316834
- DOI: 10.1101/gad.12.10.1525
Spalten, a protein containing Galpha-protein-like and PP2C domains, is essential for cell-type differentiation in Dictyostelium
Abstract
We have identified a novel gene, Spalten (Spn) that is essential for Dictyostelium multicellular development. Spn encodes a protein with an amino-terminal domain that shows very high homology to Galpha-protein subunits, a highly charged inter-region, and a carboxy-terminal domain that encodes a functional PP2C. Spn is essential for development past the mound stage, being required cell autonomously for prestalk gene expression and nonautonomously for prespore cell differentiation. Mutational analysis demonstrates that the PP2C domain is the Spn effector domain and is essential for Spn function, whereas the Galpha-like domain is required for membrane targeting and regulation of Spn function. Moreover, Spn carrying mutations in the Galpha-like domain that do not affect membrane targeting but affect specificity of guanine nucleotide binding in known GTP-binding proteins are unable to fully complement the spn- phenotype, suggesting that the Galpha-like domain regulates Spn function either directly or indirectly by mediating its interactions with other proteins. Our results suggest that Spn encodes a signaling molecule with a novel Galpha-like regulatory domain.
Figures


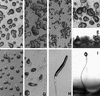



Similar articles
-
The novel ankyrin-repeat containing kinase ARCK-1 acts as a suppressor of the Spalten signaling pathway during Dictyostelium development.Dev Biol. 2003 Nov 15;263(2):308-22. doi: 10.1016/j.ydbio.2003.07.012. Dev Biol. 2003. PMID: 14597204
-
LagC is required for cell-cell interactions that are essential for cell-type differentiation in Dictyostelium.Genes Dev. 1994 Apr 15;8(8):948-58. doi: 10.1101/gad.8.8.948. Genes Dev. 1994. PMID: 7926779
-
A regulator of G protein signaling-containing kinase is important for chemotaxis and multicellular development in dictyostelium.Mol Biol Cell. 2003 Apr;14(4):1727-43. doi: 10.1091/mbc.e02-08-0550. Mol Biol Cell. 2003. PMID: 12686622 Free PMC article.
-
Glycogen synthase kinase and Dictyostelium development: old pathways pointing in new directions?Trends Genet. 1995 Feb;11(2):37-9. doi: 10.1016/s0168-9525(00)88989-1. Trends Genet. 1995. PMID: 7716803 Review. No abstract available.
-
Taking the plunge. Terminal differentiation in Dictyostelium.Trends Genet. 1999 Jan;15(1):15-9. doi: 10.1016/s0168-9525(98)01635-7. Trends Genet. 1999. PMID: 10087928 Review.
Cited by
-
Regulated protein degradation controls PKA function and cell-type differentiation in Dictyostelium.Genes Dev. 2001 Jun 1;15(11):1435-48. doi: 10.1101/gad.871101. Genes Dev. 2001. PMID: 11390363 Free PMC article.
-
Dictyostelium discoideum protein phosphatase-1 catalytic subunit exhibits distinct biochemical properties.Biochem J. 2003 Aug 1;373(Pt 3):703-11. doi: 10.1042/BJ20021964. Biochem J. 2003. PMID: 12737629 Free PMC article.
-
Purification, kinetic properties, and intracellular concentration of SpoIIE, an integral membrane protein that regulates sporulation in Bacillus subtilis.J Bacteriol. 1999 May;181(10):3242-5. doi: 10.1128/JB.181.10.3242-3245.1999. J Bacteriol. 1999. PMID: 10322028 Free PMC article.
-
Evaluation of the kinetic properties of the sporulation protein SpoIIE of Bacillus subtilis by inclusion in a model membrane.J Bacteriol. 2004 May;186(10):3195-201. doi: 10.1128/JB.186.10.3195-3201.2004. J Bacteriol. 2004. PMID: 15126482 Free PMC article.
-
Arabidopsis thaliana 'extra-large GTP-binding protein' (AtXLG1): a new class of G-protein.Plant Mol Biol. 1999 May;40(1):55-64. doi: 10.1023/a:1026483823176. Plant Mol Biol. 1999. PMID: 10394945
References
-
- Abe K, Yanagisawa K. A new class of rapid developing mutants in Dictyostelium discoideum: Implications for cyclic AMP metabolism and cell differentiation. Dev Biol. 1983;95:200–210. - PubMed
-
- Anjard C, Zeng C, Loomis WF, Nellen W. Signal transduction pathways leading to spore differentiation in Dictyostelium discoideum. Dev Biol. 1998;193:146–155. - PubMed
-
- Barford D. Molecular mechanisms of the serine/threonine phosphatases. Trends Biochem Sci. 1996;21:407–412. - PubMed
-
- Bourne HR, Sander DA, McCormick F. The GTPase superfamily: A conserved switch for diverse cell functions. Nature. 1990;348:125–132. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
