Translating old drugs into new treatments: ribosomal frameshifting as a target for antiviral agents
- PMID: 9586242
- PMCID: PMC7127214
- DOI: 10.1016/s0167-7799(97)01167-0
Translating old drugs into new treatments: ribosomal frameshifting as a target for antiviral agents
Abstract
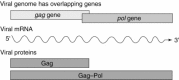
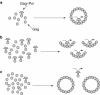
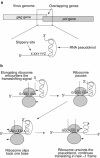
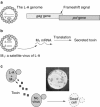
Programmed ribosomal frameshifting is used by many viruses to regulate the production of structural and enzymatic proteins. Altering the frameshifting efficiencies disrupts the virus life cycle and eliminates or reduces virus production. Ribosomal frameshifting therefore provides a unique target on which antiviral agents can function. This article describes a series of rapid assay strategies that have been developed and used to identify potential antiviral agents that target programmed -1 ribosomal frameshifting.
Figures
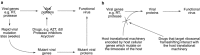

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

