Sequencing, disruption, and characterization of the Candida albicans sterol methyltransferase (ERG6) gene: drug susceptibility studies in erg6 mutants
- PMID: 9593144
- PMCID: PMC105764
- DOI: 10.1128/AAC.42.5.1160
Sequencing, disruption, and characterization of the Candida albicans sterol methyltransferase (ERG6) gene: drug susceptibility studies in erg6 mutants
Abstract
The rise in the frequency of fungal infections and the increased resistance noted to the widely employed azole antifungals make the development of new antifungals imperative for human health. The sterol biosynthetic pathway has been exploited for the development of several antifungal agents (allylamines, morpholines, azoles), but additional potential sites for antifungal agent development are yet to be fully investigated. The sterol methyltransferase gene (ERG6) catalyzes a biosynthetic step not found in humans and has been shown to result in several compromised phenotypes, most notably markedly increased permeability, when disrupted in Saccharomyces cerevisiae. The Candida albicans ERG6 gene was isolated by complementation of a S. cerevisiae erg6 mutant by using a C. albicans genomic library. Sequencing of the Candida ERG6 gene revealed high homology with the Saccharomyces version of ERG6. The first copy of the Candida ERG6 gene was disrupted by transforming with the URA3 blaster system, and the second copy was disrupted by both URA3 blaster transformation and mitotic recombination. The resulting erg6 strains were shown to be hypersusceptible to a number of sterol synthesis and metabolic inhibitors, including terbinafine, tridemorph, fenpropiomorph, fluphenazine, cycloheximide, cerulenin, and brefeldin A. No increase in susceptibility to azoles was noted. Inhibitors of the ERG6 gene product would make the cell increasingly susceptible to antifungal agents as well as to new agents which normally would be excluded and would allow for clinical treatment at lower dosages. In addition, the availability of ERG6 would allow for its use as a screen for new antifungals targeted specifically to the sterol methyltransferase.
Figures
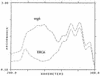



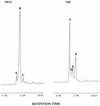
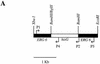


References
-
- Ausubel F, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Short protocols in molecular biology. 3rd ed. New York, N.Y: John Wiley & Sons, Inc.; 1995.
-
- Baloch R I, Mercer E I, Wiggins T E, Baldwin B C. Inhibition of ergosterol biosynthesis in Saccharomyces cerevisiae and Ustilago maydis by tridemorph, fenpropiomorph, and fenpropidin. Phytochemistry. 1984;23:2219–2226.
-
- Barrett-Bee K, Dixon G. Ergosterol biosynthesis inhibition: a target for antifungal agents. Acta Biochim Pol. 1995;42:465–480. - PubMed
-
- Boeke J D, Truehart J, Natsoulis G, Fink G R. 5-Fluoro-orotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

