Modification of Ras in eukaryotic cells by Pseudomonas aeruginosa exoenzyme S
- PMID: 9596723
- PMCID: PMC108245
- DOI: 10.1128/IAI.66.6.2607-2613.1998
Modification of Ras in eukaryotic cells by Pseudomonas aeruginosa exoenzyme S
Abstract
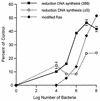
Genetic and functional data suggest that Pseudomonas aeruginosa exoenzyme S (ExoS), an ADP-ribosyltransferase, is translocated into eukaryotic cells by a bacterial type III secretory mechanism activated by contact between bacteria and host cells. Although purified ExoS is not toxic to eukaryotic cells, ExoS-producing bacteria cause reduced proliferation and viability, possibly mediated by bacterially translocated ExoS. To investigate the activity of translocated ExoS, we examined in vivo modification of Ras, a preferred in vitro substrate. The ExoS-producing strain P. aeruginosa 388 and an isogenic mutant strain, 388DeltaexoS, which fails to produce ExoS, were cocultured with HT29 colon carcinoma cells. Ras was found to be ADP-ribosylated during coculture with 388 but not with 388DeltaexoS, and Ras modification by 388 corresponded with reduction in HT29 cell DNA synthesis. Active translocation by bacteria was found to be required, since exogenous ExoS, alone or in the presence of 388DeltaexoS, was unable to modify intracellular Ras. Other ExoS-producing strains caused modification of Ras, indicating that this is not a strain-specific event. ADP-ribosylation of Rap1, an additional Ras family substrate for ExoS in vitro, was not detectable in vivo under conditions sufficient for Ras modification, suggesting possible ExoS substrate preference among Ras-related proteins. These results confirm that intracellular Ras is modified by bacterially translocated ExoS and that the inhibition of target cell proliferation correlates with the efficiency of Ras modification.
Figures



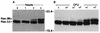

Similar articles
-
Independent and coordinate effects of ADP-ribosyltransferase and GTPase-activating activities of exoenzyme S on HT-29 epithelial cell function.Infect Immun. 2001 Sep;69(9):5318-28. doi: 10.1128/IAI.69.9.5318-5328.2001. Infect Immun. 2001. PMID: 11500401 Free PMC article.
-
Pseudomonas aeruginosa exoenzyme S ADP-ribosylates Ras at multiple sites.J Biol Chem. 1998 Mar 27;273(13):7332-7. doi: 10.1074/jbc.273.13.7332. J Biol Chem. 1998. PMID: 9516428
-
ADP-ribosylation of oncogenic Ras proteins by pseudomonas aeruginosa exoenzyme S in vivo.Mol Microbiol. 1999 Jun;32(5):1054-64. doi: 10.1046/j.1365-2958.1999.01420.x. Mol Microbiol. 1999. PMID: 10361307
-
Pseudomonas aeruginosa exoenzyme S, a bifunctional type-III secreted cytotoxin.Int J Med Microbiol. 2000 Oct;290(4-5):381-7. doi: 10.1016/S1438-4221(00)80047-8. Int J Med Microbiol. 2000. PMID: 11111915 Review.
-
The exoenzyme S regulon of Pseudomonas aeruginosa.Mol Microbiol. 1997 Nov;26(4):621-9. doi: 10.1046/j.1365-2958.1997.6251991.x. Mol Microbiol. 1997. PMID: 9427393 Review.
Cited by
-
Multiple domains are required for the toxic activity of Pseudomonas aeruginosa ExoU.J Bacteriol. 2001 Jul;183(14):4330-44. doi: 10.1128/JB.183.14.4330-4344.2001. J Bacteriol. 2001. PMID: 11418575 Free PMC article.
-
The impact of simvastatin on pulmonary effectors of Pseudomonas aeruginosa infection.PLoS One. 2014 Jul 10;9(7):e102200. doi: 10.1371/journal.pone.0102200. eCollection 2014. PLoS One. 2014. PMID: 25010049 Free PMC article.
-
Independent and coordinate effects of ADP-ribosyltransferase and GTPase-activating activities of exoenzyme S on HT-29 epithelial cell function.Infect Immun. 2001 Sep;69(9):5318-28. doi: 10.1128/IAI.69.9.5318-5328.2001. Infect Immun. 2001. PMID: 11500401 Free PMC article.
-
Exotoxin S secreted by internalized Pseudomonas aeruginosa delays lytic host cell death.PLoS Pathog. 2022 Feb 7;18(2):e1010306. doi: 10.1371/journal.ppat.1010306. eCollection 2022 Feb. PLoS Pathog. 2022. PMID: 35130333 Free PMC article.
-
Sera from adult patients with cystic fibrosis contain antibodies to Pseudomonas aeruginosa type III apparatus.Infect Immun. 2001 Feb;69(2):1185-8. doi: 10.1128/IAI.69.2.1185-1188.2001. Infect Immun. 2001. PMID: 11160019 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

