Dynamic regulation of copper uptake and detoxification genes in Saccharomyces cerevisiae
- PMID: 9599102
- PMCID: PMC110631
- DOI: 10.1128/MCB.18.5.2514
Dynamic regulation of copper uptake and detoxification genes in Saccharomyces cerevisiae
Abstract
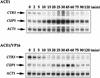
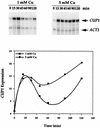
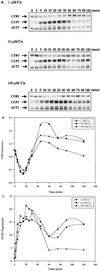
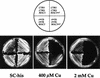
The essential yet toxic nature of copper demands tight regulation of the copper homeostatic machinery to ensure that sufficient copper is present in the cell to drive essential biochemical processes yet prevent the accumulation to toxic levels. In Saccharomyces cerevisiae, the nutritional copper sensor Mac1p regulates the copper-dependent expression of the high affinity Cu(I) uptake genes CTR1, CTR3, and FRE1, while the toxic copper sensor Ace1p regulates the transcriptional activation of the detoxification genes CUP1, CRS5, and SOD1 in response to copper. In this study, we characterized the tandem regulation of the copper uptake and detoxification pathways in response to the chronic presence of elevated concentrations of copper ions in the growth medium. Upon addition of CuSO4, mRNA levels of CTR3 were rapidly reduced to eightfold the original basal level whereas the Ace1p-mediated transcriptional activation of CUP1 was rapid and potent but transient. CUP1 expression driven by an Ace1p DNA binding domain-herpes simplex virus VP16 transactivation domain fusion was also transient, demonstrating that this mode of regulation occurs via modulation of the Ace1p copper-activated DNA binding domain. In vivo dimethyl sulfate footprinting analysis of the CUP1 promoter demonstrated transient occupation of the metal response elements by Ace1p which paralleled CUP1 mRNA expression. Analysis of a Mac1p mutant, refractile for copper-dependent repression of the Cu(I) transport genes, showed an aberrant pattern of CUP1 expression and copper sensitivity. These studies (i) demonstrate that the nutritional and toxic copper metalloregulatory transcription factors Mac1p and Ace1p must sense and respond to copper ions in a dynamic fashion to appropriately regulate copper ion homeostasis and (ii) establish the requirement for a wild-type Mac1p for survival in the presence of toxic copper levels.
Figures




References
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1987.
-
- Bull P C, Cox D W. Wilson disease and Menkes disease: new handles on heavy metal transport. Trends Genet Sci. 1994;10:248–252. - PubMed
-
- Bull P C, Thomas G R, Rommens J M, Forbe J R, Cox D W. The Wilson disease gene is a putative copper transporting P-type ATPase similar to the Menkes gene. Nat Genet. 1993;5:327–337. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
