Photoactive yellow protein: a structural prototype for the three-dimensional fold of the PAS domain superfamily
- PMID: 9600888
- PMCID: PMC34491
- DOI: 10.1073/pnas.95.11.5884
Photoactive yellow protein: a structural prototype for the three-dimensional fold of the PAS domain superfamily
Abstract
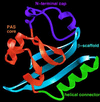
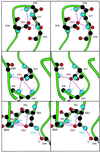
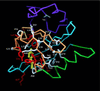
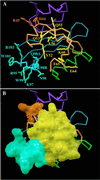
PAS domains are found in diverse proteins throughout all three kingdoms of life, where they apparently function in sensing and signal transduction. Although a wealth of useful sequence and functional information has become recently available, these data have not been integrated into a three-dimensional (3D) framework. The very early evolutionary development and diverse functions of PAS domains have made sequence analysis and modeling of this protein superfamily challenging. Limited sequence similarities between the approximately 50-residue PAS repeats and one region of the bacterial blue-light photosensor photoactive yellow protein (PYP), for which ground-state and light-activated crystallographic structures have been determined to high resolution, originally were identified in sequence searches using consensus sequence probes from PAS-containing proteins. Here, we found that by changing a few residues particular to PYP function, the modified PYP sequence probe also could select PAS protein sequences. By mapping a typical approximately 150-residue PAS domain sequence onto the entire crystallographic structure of PYP, we show that the PAS sequence similarities and differences are consistent with a shared 3D fold (the PAS/PYP module) with obvious potential for a ligand-binding cavity. Thus, PYP appears to prototypically exhibit all the major structural and functional features characteristic of the PAS domain superfamily: the shared PAS/PYP modular domain fold of approximately 125-150 residues, a sensor function often linked to ligand or cofactor (chromophore) binding, and signal transduction capability governed by heterodimeric assembly (to the downstream partner of PYP). This 3D PAS/PYP module provides a structural model to guide experimental testing of hypotheses regarding ligand-binding, dimerization, and signal transduction.
Figures

References
-
- Nambu J R, Lewis J O, Jr, Wharton K A, Crews S T. Cell. 1991;67:1157–1167. - PubMed
-
- Crews S T, Thomas J B, Goodman C S. Cell. 1988;52:143–151. - PubMed
-
- Hoffman E C, Reyes H, Chu F-F, Sander F, Conley L H, Brooks B A, Hankinson O. Science. 1991;252:954–958. - PubMed
-
- Ponting C P, Aravind L. Curr Biol. 1997;7:674–677. - PubMed
-
- Zhulin I B, Taylor B L, Dixon R. Trends Biochem Sci. 1997;22:331–333. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

