Submillimolar levels of calcium regulates DNA structure at the dinucleotide repeat (TG/AC)n
- PMID: 9600903
- PMCID: PMC27571
- DOI: 10.1073/pnas.95.11.5981
Submillimolar levels of calcium regulates DNA structure at the dinucleotide repeat (TG/AC)n
Abstract
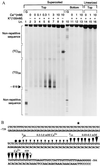
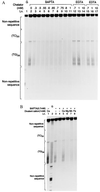
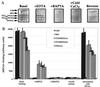
Submillimolar levels of calcium, similar to the physiological total (bound + free) intranuclear concentration (0.01-1 mM), induced a conformational change within d(TG/AC)n, one of the frequent dinucleotide repeats of the mammalian genome. This change is calcium-specific, because no other tested cation induced it and it was detected as a concentration-dependent transition from B- to a non-B-DNA conformation expanding from 3' end toward the 5' of the repeat. Genomic footprinting of various rat brain regions revealed the existence of similar non-B-DNA conformation within a d(TG/AC)28 repeat of the endogenous enkephalin gene only in enkephalin-expressing caudate nucleus and not in the nonexpressing thalamus. Binding assays demonstrated that DNA could bind calcium and can compete with calmodulin for calcium.
Figures


Similar articles
-
Isolation and structural and genetic analysis of the mouse enkephalin gene and its d(AC/TG)n repeats.DNA Seq. 1998;9(4):217-26. doi: 10.3109/10425179809105208. DNA Seq. 1998. PMID: 10520752
-
A novel rat genomic simple repeat DNA with RNA-homology shows triplex (H-DNA)-like structure and tissue-specific RNA expression.Biochem Biophys Res Commun. 2005 Feb 4;327(1):276-86. doi: 10.1016/j.bbrc.2004.12.015. Biochem Biophys Res Commun. 2005. PMID: 15629459
-
Perturbations in DNA structure upon interaction with porphyrins revealed by chemical probes, DNA footprinting and molecular modelling.Bioorg Med Chem. 1995 Jun;3(6):671-7. doi: 10.1016/0968-0896(95)00052-i. Bioorg Med Chem. 1995. PMID: 7582945
-
Topoisomerase II-mediated DNA cleavage on the cruciform structure formed within the 5'upstream region of the human beta-globin gene.Mol Cells. 1998 Aug 31;8(4):424-30. Mol Cells. 1998. PMID: 9749529
-
The non-B-DNA structure of d(CA/TG)n does not differ from that of Z-DNA.Proc Natl Acad Sci U S A. 1994 Sep 27;91(20):9549-53. doi: 10.1073/pnas.91.20.9549. Proc Natl Acad Sci U S A. 1994. PMID: 7937803 Free PMC article.
Cited by
-
Power-law rheology of isolated nuclei with deformation mapping of nuclear substructures.Biophys J. 2005 Oct;89(4):2855-64. doi: 10.1529/biophysj.105.062554. Epub 2005 Jul 29. Biophys J. 2005. PMID: 16055543 Free PMC article.
-
The Role of Lamins in the Nucleoplasmic Reticulum, a Pleiomorphic Organelle That Enhances Nucleo-Cytoplasmic Interplay.Front Cell Dev Biol. 2022 Jun 16;10:914286. doi: 10.3389/fcell.2022.914286. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35784476 Free PMC article. Review.
-
Regulation of calcium signals in the nucleus by a nucleoplasmic reticulum.Nat Cell Biol. 2003 May;5(5):440-6. doi: 10.1038/ncb980. Nat Cell Biol. 2003. PMID: 12717445 Free PMC article.
-
The interplay of transcription factors in suppression of UV-B induced flavonol accumulation by flg22.Plant Signal Behav. 2014 Apr 10;9(4):e28745. doi: 10.4161/psb.28745. Online ahead of print. Plant Signal Behav. 2014. PMID: 24721804 Free PMC article.
-
Nucleoplasmic calcium signaling and cell proliferation: calcium signaling in the nucleus.Cell Commun Signal. 2013 Feb 21;11(1):14. doi: 10.1186/1478-811X-11-14. Cell Commun Signal. 2013. PMID: 23433362 Free PMC article.
References
-
- Tvedt K E, Kopstad G, Haugen O A, Halgunset J. Cancer Res. 1987;47:323–328. - PubMed
-
- Ornberg R L, Kuijpers G A, Leapman R D. J Biol Chem. 1988;263:1488–1493. - PubMed
-
- Chandra S, Fewtrell C, Millard P J, Sandison D R, Webb W W, Morrison G H. J Biol Chem. 1994;269:15186–15194. - PubMed
-
- Bock G R, Ackrill K. Calcium Waves, Gradients and Oscillations, CIBA Found. Symp. Vol. 188. Chichester, U.K.: Wiley; 1995. pp. 1–281. - PubMed
-
- Bootman M D, Berridge M J. Cell. 1995;83:675–678. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

