Crystallographic comparison of the estrogen and progesterone receptor's ligand binding domains
- PMID: 9600906
- PMCID: PMC27574
- DOI: 10.1073/pnas.95.11.5998
Crystallographic comparison of the estrogen and progesterone receptor's ligand binding domains
Abstract
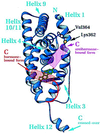
The 2.8-A crystal structure of the complex formed by estradiol and the human estrogen receptor-alpha ligand binding domain (hERalphaLBD) is described and compared with the recently reported structure of the progesterone complex of the human progesterone receptor ligand binding domain, as well as with similar structures of steroid/nuclear receptor LBDs solved elsewhere. The hormone-bound hERalphaLBD forms a distinctly different and probably more physiologically important dimer interface than its progesterone counterpart. A comparison of the specificity determinants of hormone binding reveals a common structural theme of mutually supported van der Waals and hydrogen-bonded interactions involving highly conserved residues. The previously suggested mechanism by which the estrogen receptor distinguishes estradiol's unique 3-hydroxy group from the 3-keto function of most other steroids is now described in atomic detail. Mapping of mutagenesis results points to a coactivator-binding surface that includes the region around the "signature sequence" as well as helix 12, where the ligand-dependent conformation of the activation function 2 core is similar in all previously solved steroid/nuclear receptor LBDs. A peculiar crystal packing event displaces helix 12 in the hERalphaLBD reported here, suggesting a higher degree of dynamic variability than expected for this critical substructure.
Figures



Similar articles
-
Atomic structure of progesterone complexed with its receptor.Nature. 1998 May 28;393(6683):392-6. doi: 10.1038/30775. Nature. 1998. PMID: 9620806
-
The conserved residues of the ligand-binding domains of steroid receptors are located in the core of the molecules.J Mol Graph Model. 2001;19(6):543-51, 601-6. doi: 10.1016/s1093-3263(01)00087-0. J Mol Graph Model. 2001. PMID: 11552682
-
Estrogen and progesterone receptors: from molecular structures to clinical targets.Cell Mol Life Sci. 2009 Aug;66(15):2405-26. doi: 10.1007/s00018-009-0017-3. Epub 2009 Mar 31. Cell Mol Life Sci. 2009. PMID: 19333551 Free PMC article. Review.
-
Electrostatic interactions of androgens and progesterone derivatives with rainbow trout estrogen receptor.J Steroid Biochem Mol Biol. 2000 Dec 15;75(2-3):129-37. doi: 10.1016/s0960-0760(00)00162-x. J Steroid Biochem Mol Biol. 2000. PMID: 11226829
-
The significance of the 20-carbonyl group of progesterone in steroid receptor binding: a molecular dynamics and structure-based ligand design study.Steroids. 2003 Nov;68(10-13):869-78. doi: 10.1016/j.steroids.2003.08.009. Steroids. 2003. PMID: 14667979 Review.
Cited by
-
Blockade of the Hedgehog pathway downregulates estrogen receptor alpha signaling in breast cancer cells.Oncotarget. 2016 Nov 1;7(44):71580-71593. doi: 10.18632/oncotarget.12259. Oncotarget. 2016. PMID: 27689403 Free PMC article.
-
Random multi-recombinant PCR for the construction of combinatorial protein libraries.Nucleic Acids Res. 2001 Oct 15;29(20):E97. doi: 10.1093/nar/29.20.e97. Nucleic Acids Res. 2001. PMID: 11600716 Free PMC article.
-
Retinoid isomers differ in the ability to induce release of SMRT corepressor from retinoic acid receptor-alpha.J Biol Chem. 1999 Jan 29;274(5):2885-92. doi: 10.1074/jbc.274.5.2885. J Biol Chem. 1999. PMID: 9915825 Free PMC article.
-
In vivo effects of a GPR30 antagonist.Nat Chem Biol. 2009 Jun;5(6):421-7. doi: 10.1038/nchembio.168. Nat Chem Biol. 2009. PMID: 19430488 Free PMC article.
-
Synthesis and evaluation of 17α-E-20-(heteroaryl)norpregn-1,3,5(10),20 tetraene-3,17β-diols [17α-(heteroaryl)vinyl estradiols] as ligands for the estrogen receptor-α ligand binding domain (ERα-LBD).Bioorg Med Chem Lett. 2012 Jan 15;22(2):977-9. doi: 10.1016/j.bmcl.2011.12.003. Epub 2011 Dec 8. Bioorg Med Chem Lett. 2012. PMID: 22178552 Free PMC article.
References
-
- Brzozowski A M, Pike A C W, Dauter Z, Hubbard R E, Bonn T, Engström O, Öhman L, Greene G L, Gustafsson J, Carlquist M. Nature (London) 1997;389:753–758. - PubMed
-
- Wurtz J M, Bourguet W, Renaud J P, Vivat V, Chambon P, Moras D, Gronemeyer H. Nat Struct Biol. 1996;3:87–94. - PubMed
-
- Katzenellenbogen J A, Katzenellenbogen B S. Chem Biol. 1996;3:529–536. - PubMed
-
- Thénot S, Henriquet C, Rochefort H, Cavaillès V. J Biol Chem. 1997;272:12062–12068. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

