Identification of an additional negative regulatory region for p53 sequence-specific DNA binding
- PMID: 9600920
- PMCID: PMC27588
- DOI: 10.1073/pnas.95.11.6079
Identification of an additional negative regulatory region for p53 sequence-specific DNA binding
Abstract
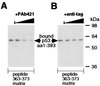
The DNA binding activity of p53 is crucial for its tumor suppressor function and is subject to tight regulation. Previous studies revealed that the inhibitory function of the p53 C terminus is implicated in the latent, low affinity sequence-specific DNA binding activity of p53 in the uninduced state. Sequence-specific DNA binding of p53 has been shown to be activated by several posttranslational modifications and interacting proteins that target predominantly the C terminus. Moreover, several authors have shown that synthetic peptides corresponding to p53 C-terminal sequences activate p53 sequence-specific DNA binding. In an effort to identify the interaction site of p53 with these activating peptides we assessed complex formation between p53 deletion constructs and C-terminal activating peptides by peptide affinity precipitation. This study revealed that two distal regions of the p53 molecule contribute synergistically to the interaction with activating C-terminal peptides: amino acids 80-93 and 364-393. The C-terminal residues 364-393 are already well characterized as having negative regulatory function. DNA binding analyses with these deletion constructs reveal a comparable negative regulatory activity for residues 80-93, defining this region as a previously unidentified negative regulatory domain of p53. Furthermore, synthetic peptides spanning this newly identified proline-rich negative regulatory region (residues 80-93) are able to activate p53 sequence-specific DNA binding in vitro. We suggest that both negative regulatory regions, residues 80-93 and 364-393, contribute cooperatively to the maintenance of the latent, low-affinity DNA binding conformation of p53.
Figures


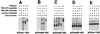


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

