Generation of broad-spectrum disease resistance by overexpression of an essential regulatory gene in systemic acquired resistance
- PMID: 9601001
- PMCID: PMC34547
- DOI: 10.1073/pnas.95.11.6531
Generation of broad-spectrum disease resistance by overexpression of an essential regulatory gene in systemic acquired resistance
Abstract
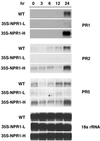
The recently cloned NPR1 gene of Arabidopsis thaliana is a key regulator of acquired resistance responses. Upon induction, NPR1 expression is elevated and the NPR1 protein is activated, in turn inducing expression of a battery of downstream pathogenesis-related genes. In this study, we found that NPR1 confers resistance to the pathogens Pseudomonas syringae and Peronospora parasitica in a dosage-dependent fashion. Overexpression of NPR1 leads to enhanced resistance with no obvious detrimental effect on the plants. Thus, for the first time, a single gene is shown to be a workable target for genetic engineering of nonspecific resistance in plants.
Figures



References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

