Identification and characterization of MAE1, the Saccharomyces cerevisiae structural gene encoding mitochondrial malic enzyme
- PMID: 9603875
- PMCID: PMC107252
- DOI: 10.1128/JB.180.11.2875-2882.1998
Identification and characterization of MAE1, the Saccharomyces cerevisiae structural gene encoding mitochondrial malic enzyme
Abstract
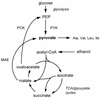
Pyruvate, a precursor for several amino acids, can be synthesized from phosphoenolpyruvate by pyruvate kinase. Nevertheless, pyk1 pyk2 mutants of Saccharomyces cerevisiae devoid of pyruvate kinase activity grew normally on ethanol in defined media, indicating the presence of an alternative route for pyruvate synthesis. A candidate for this role is malic enzyme, which catalyzes the oxidative decarboxylation of malate to pyruvate. Disruption of open reading frame YKL029c, which is homologous to malic enzyme genes from other organisms, abolished malic enzyme activity in extracts of glucose-grown cells. Conversely, overexpression of YKL029c/MAE1 from the MET25 promoter resulted in an up to 33-fold increase of malic enzyme activity. Growth studies with mutants demonstrated that presence of either Pyk1p or Mae1p is required for growth on ethanol. Mutants lacking both enzymes could be rescued by addition of alanine or pyruvate to ethanol cultures. Disruption of MAE1 alone did not result in a clear phenotype. Regulation of MAE1 was studied by determining enzyme activities and MAE1 mRNA levels in wild-type cultures and by measuring beta-galactosidase activities in a strain carrying a MAE1::lacZ fusion. Both in shake flask cultures and in carbon-limited chemostat cultures, MAE1 was constitutively expressed. A three- to fourfold induction was observed during anaerobic growth on glucose. Subcellular fractionation experiments indicated that malic enzyme in S. cerevisiae is a mitochondrial enzyme. Its regulation and localization suggest a role in the provision of intramitochondrial NADPH or pyruvate under anaerobic growth conditions. However, since null mutants could still grow anaerobically, this function is apparently not essential.
Figures

References
-
- Boles E. Pyruvate kinase. In: Entian K-D, Zimmermann F K, editors. Yeast sugar metabolism: biochemistry, genetics, biotechnology and applications. Lancaster, Pa: Technomic Publishing Co.; 1997. pp. 171–186.
-
- Boles, E. Unpublished data.
-
- Boles E, Schulte F, Miosga T, Freidel K, Schlüter E, Zimmermann F K, Hollenberg C P, Heinisch J J. Characterization of a glucose-repressed pyruvate kinase (Pyk2p) in Saccharomyces cerevisiae that is catalytically insensitive to fructose-1,6-bisphosphate. J Bacteriol. 1997;179:2987–2993. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

