Posttranscriptional modifications in 16S and 23S rRNAs of the archaeal hyperthermophile Sulfolobus solfataricus
- PMID: 9603876
- PMCID: PMC107253
- DOI: 10.1128/JB.180.11.2883-2888.1998
Posttranscriptional modifications in 16S and 23S rRNAs of the archaeal hyperthermophile Sulfolobus solfataricus
Abstract
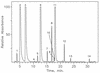
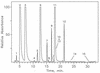
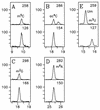
Posttranscriptional modification is common to many types of RNA, but the majority of information concerning structure and function of modification is derived principally from tRNA. By contrast, less is known about modification in rRNA in spite of accumulating evidence for its direct participation in translation. The structural identities and approximate molar levels of modifications have been established for 16S and 23S rRNAs of the archaeal hyperthermophile Sulfolobus solfactaricus by using combined chromatography-mass spectrometry-based methods. Modification levels are exceptionally high for prokaryotic organisms, with approximately 38 modified sites in 16S rRNA and 50 in 23S rRNA for cells cultured at 75 degrees C, compared with 11 and 23 sites, respectively, in Escherichia coli. We structurally characterized 10 different modified nucleosides in 16S rRNA, 64% (24 residues) of which are methylated at O-2' of ribose, and 8 modified species in 23S rRNA, 86% (43 residues) of which are ribose methylated, a form of modification shown in earlier studies to enhance stability of the polynucleotide chain. From cultures grown at progressively higher temperatures, 60, 75, and 83 degrees C, a slight trend toward increased ribose methylation levels was observed, with greatest net changes over the 23 degrees C range shown for 2'-O-methyladenosine in 16S rRNA (21% increase) and for 2'-O-methylcytidine (24%) and 2'-O-methylguanosine (22%) in 23S rRNA. These findings are discussed in terms of the potential role of modification in stabilization of rRNA in the thermal environment.
Figures
References
-
- Agris P F, Koh P, Söll D. The effect of growth temperature on the in vivo ribose methylation of Bacillus stearothermophilus. Arch Biochem Biophys. 1973;154:277–282. - PubMed
-
- Björk G, Ericson J U, Gustafsson C E D, Hagervall T G, Jönsson Y H, Wikström P M. Transfer RNA modification. Annu Rev Biochem. 1987;56:263–287. - PubMed
-
- Björk G. The role of modified nucleosides in tRNA interactions. In: Hatfield D L, Lee B J, Pirtle R M, editors. Transfer RNA in protein synthesis. Boca Raton, Fla: CRC Press; 1992. pp. 23–85.
-
- Brimacombe R, Mitchell P, Osswald M, Stade K, Bochkariov D. Clustering of modified nucleotides at the functional center of bacterial ribosomal RNA. FASEB J. 1993;7:161–167. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

