Lumican regulates collagen fibril assembly: skin fragility and corneal opacity in the absence of lumican
- PMID: 9606218
- PMCID: PMC2137175
- DOI: 10.1083/jcb.141.5.1277
Lumican regulates collagen fibril assembly: skin fragility and corneal opacity in the absence of lumican
Abstract
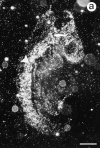
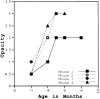
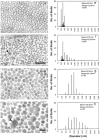
Lumican, a prototypic leucine-rich proteoglycan with keratan sulfate side chains, is a major component of the cornea, dermal, and muscle connective tissues. Mice homozygous for a null mutation in lumican display skin laxity and fragility resembling certain types of Ehlers-Danlos syndrome. In addition, the mutant mice develop bilateral corneal opacification. The underlying connective tissue defect in the homozygous mutants is deregulated growth of collagen fibrils with a significant proportion of abnormally thick collagen fibrils in the skin and cornea as indicated by transmission electron microscopy. A highly organized and regularly spaced collagen fibril matrix typical of the normal cornea is also missing in these mutant mice. This study establishes a crucial role for lumican in the regulation of collagen assembly into fibrils in various connective tissues. Most importantly, these results provide a definitive link between a necessity for lumican in the development of a highly organized collagenous matrix and corneal transparency.
Figures














References
-
- Birk DE, Fitch JM, Babiarz JP, Doane KJ, Linsenmayer TE. Collagen fibrillogenesis in vitro: interactions of types I and V collagen regulates fibril diameter. J Cell Sci. 1990;95:649–657. - PubMed
-
- Blochberger TC, Vergnes J-P, Hempel J, Hassell JR. cDNA to chick lumican (corneal keratan sulfate proteoglycan) reveals homology to the small interstitial proteoglycan gene family and expression in muscle and intestine. J Biol Chem. 1992;267:347–352. - PubMed
-
- Burch GH, Gong Y, Liu W, Dettman RW, Curry CJ, Smith L, Miller WL, Bristow J. Tenascin-X deficiency is associated with Ehlers-Danlos syndrome. Nat Genet. 1997;17:104–108. - PubMed
-
- Byers PH, Holbrook KA, Hall JG, Bornstein P, Chandler JW. A new variety of spondyoepiphyseal dysplasia characterized by punctate corneal dystrophy and abnormal collagen fibrils. Hum Genet. 1978;40:157–169. - PubMed
-
- Casey, T.A., and K.W. Sharif. 1991. A Color Atlas of Corneal Dystrophies and Degenerations. Wolfe Publishing Limited, London. 21–51.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

