Yeast carbon catabolite repression
- PMID: 9618445
- PMCID: PMC98918
- DOI: 10.1128/MMBR.62.2.334-361.1998
Yeast carbon catabolite repression
Abstract
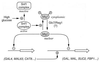
Glucose and related sugars repress the transcription of genes encoding enzymes required for the utilization of alternative carbon sources; some of these genes are also repressed by other sugars such as galactose, and the process is known as catabolite repression. The different sugars produce signals which modify the conformation of certain proteins that, in turn, directly or through a regulatory cascade affect the expression of the genes subject to catabolite repression. These genes are not all controlled by a single set of regulatory proteins, but there are different circuits of repression for different groups of genes. However, the protein kinase Snf1/Cat1 is shared by the various circuits and is therefore a central element in the regulatory process. Snf1 is not operative in the presence of glucose, and preliminary evidence suggests that Snf1 is in a dephosphorylated state under these conditions. However, the enzymes that phosphorylate and dephosphorylate Snf1 have not been identified, and it is not known how the presence of glucose may affect their activity. What has been established is that Snf1 remains active in mutants lacking either the proteins Grr1/Cat80 or Hxk2 or the Glc7 complex, which functions as a protein phosphatase. One of the main roles of Snf1 is to relieve repression by the Mig1 complex, but it is also required for the operation of transcription factors such as Adr1 and possibly other factors that are still unidentified. Although our knowledge of catabolite repression is still very incomplete, it is possible in certain cases to propose a partial model of the way in which the different elements involved in catabolite repression may be integrated.
Figures
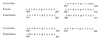
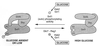
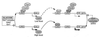
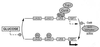
References
-
- Altamura N, Dujardin G, Groudinsky O, Slonimski P P. Two adjacent nuclear genes, ISF1 and NAM7/UPF1, cooperatively participate in mitochondrial functions in Saccharomyces cerevisiae. Mol Gen Genet. 1994;242:49–56. - PubMed
-
- Bailey R B, Woodward A. Isolation and characterization of a pleiotropic glucose repression resistant mutant of Saccharomyces cerevisiae. Mol Gen Genet. 1984;193:507–512. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

