Antiviral agent blocks breathing of the common cold virus
- PMID: 9618488
- PMCID: PMC22631
- DOI: 10.1073/pnas.95.12.6774
Antiviral agent blocks breathing of the common cold virus
Abstract
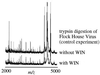
A dynamic capsid is critical to the events that shape the viral life cycle; events such as cell attachment, cell entry, and nucleic acid release demand a highly mobile viral surface. Protein mass mapping of the common cold virus, human rhinovirus 14 (HRV14), revealed both viral structural dynamics and the inhibition of such dynamics with an antiviral agent, WIN 52084. Viral capsid digestion fragments resulting from proteolytic time-course experiments provided structural information in good agreement with the HRV14 three-dimensional crystal structure. As expected, initial digestion fragments included peptides from the capsid protein VP1. This observation was expected because VP1 is the most external viral protein. Initial digestion fragments also included peptides belonging to VP4, the most internal capsid protein. The mass spectral results together with x-ray crystallography data provide information consistent with a "breathing" model of the viral capsid. Whereas the crystal structure of HRV14 shows VP4 to be the most internal capsid protein, mass spectral results show VP4 fragments to be among the first digestion fragments observed. Taken together this information demonstrates that VP4 is transiently exposed to the viral surface via viral breathing. Comparative digests of HRV14 in the presence and absence of WIN 52084 revealed a dramatic inhibition of digestion. These results indicate that the binding of the antiviral agent not only causes local conformational changes in the drug binding pocket but actually stabilizes the entire viral capsid against enzymatic degradation. Viral capsid mass mapping provides a fast and sensitive method for probing viral structural dynamics as well as providing a means for investigating antiviral drug efficacy.
Figures



References
-
- Rueckert R R. Picornaviridae and Their Replication. New York: Raven; 1996. pp. 609–654.
-
- Rossmann, M. G., Smith, T. J. & Rueckert, R. R. (1993) Structure Introductory Issue, xxiv–xxv.
-
- Kriwacki R W, Wu J, Siuzdak G, Wright P E. J Am Chem Soc. 1996;118:5320–5321.
-
- Bothner B, Dong X-F, Bibbs L, Johnson J E, Siuzdak G. J Biol Chem. 1998;273:673–676. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

