SIRE-1, a copia/Ty1-like retroelement from soybean, encodes a retroviral envelope-like protein
- PMID: 9618510
- PMCID: PMC22677
- DOI: 10.1073/pnas.95.12.6897
SIRE-1, a copia/Ty1-like retroelement from soybean, encodes a retroviral envelope-like protein
Abstract
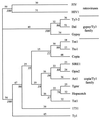
The soybean genome hosts a family of several hundred, relatively homogeneous copies of a large, copia/Ty1-like retroelement designated SIRE-1. A copy of this element has been recovered from a Glycine max genomic library. DNA sequence analysis of two SIRE-1 subclones revealed that SIRE-1 contains a long, uninterrupted, ORF between the 3' end of the pol ORF and the 3' long terminal repeat (LTR), a region that harbors the env gene in retroviral genomes. Conceptual translation of this second ORF produces a 70-kDa protein. Computer analyses of the amino acid sequence predicted patterns of transmembrane domains, alpha-helices, and coiled coils strikingly similar to those found in mammalian retroviral envelope proteins. In addition, a 65-residue, proline-rich domain is characterized by a strong amino acid compositional bias virtually identical to that of the 60-amino acid, proline-rich neutralization domain of the feline leukemia virus surface protein. The assignment of SIRE-1 to the copia/Ty1 family was confirmed by comparison of the conceptual translation of its reverse transcriptase-like domain with those of other retroelements. This finding suggests the presence of a proretrovirus in a plant genome and is the strongest evidence to date for the existence of a retrovirus-like genome closely related to copia/Ty1 retrotransposons.
Figures

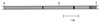
 , cDNA overlap; ▩,
flanking ORF. H, HindIII; X, XbaI.
, cDNA overlap; ▩,
flanking ORF. H, HindIII; X, XbaI.


References
-
- Eickbush T H. In: The Evolutionary Biology of Viruses. Morse S S, editor. New York: Raven; 1994. pp. 121–157.
-
- Varmus H, Brown P. In: Mobile DNA. Berg D E, Howe M M, editors. Washington, DC: Am. Soc. Microbiol.; 1989. pp. 53–108.
-
- Flavell A J. Comp Biochem Physiol B. 1995;110:3–15. - PubMed
-
- Flavell A J, Jackson V, Iqbal M P, Riach I, Waddell S. Mol Gen Genet. 1995;246:65–71. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

