Assessment of resolution and intercenter reproducibility of results of genotyping Staphylococcus aureus by pulsed-field gel electrophoresis of SmaI macrorestriction fragments: a multicenter study
- PMID: 9620395
- PMCID: PMC104895
- DOI: 10.1128/JCM.36.6.1653-1659.1998
Assessment of resolution and intercenter reproducibility of results of genotyping Staphylococcus aureus by pulsed-field gel electrophoresis of SmaI macrorestriction fragments: a multicenter study
Abstract
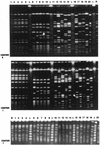
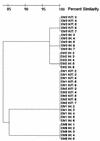
Twenty well-characterized isolates of methicillin-resistant Staphylococcus aureus were used to study the optimal resolution and interlaboratory reproducibility of pulsed-field gel electrophoresis (PFGE) of DNA macrorestriction fragments. Five identical isolates (one PFGE type), 5 isolates that produced related PFGE subtypes, and 10 isolates with unique PFGE patterns were analyzed blindly in 12 different laboratories by in-house protocols. In several laboratories a standardized PFGE protocol with a commercial kit was applied successfully as well. Eight of the centers correctly identified the genetic homogeneity of the identical isolates by both the in-house and standard protocols. Four of 12 laboratories failed to produce interpretable data by the standardized protocol, due to technical problems (primarily plug preparation). With the five related isolates, five of eight participants identified the same subtype interrelationships with both in-house and standard protocols. However, two participants identified multiple strain types in this group or classified some of the isolates as unrelated isolates rather than as subtypes. The remaining laboratory failed to distinguish differences between some of the related isolates by utilizing both the in-house and standardized protocols. There were large differences in the relative genome lengths of the isolates as calculated on the basis of the gel pictures. By visual inspection, the numbers of restriction fragments and overall banding pattern similarity in the three groups of isolates showed interlaboratory concordance, but centralized computer analysis of data from four laboratories yielded percent similarity values of only 85% for the group of identical isolates. The differences between the data sets obtained with in-house and standardized protocols could be the experimental parameters which differed with respect to the brand of equipment used, imaging software, running time (20 to 48 h), and pulsing conditions. In conclusion, it appears that the standardization of PFGE depends on controlling a variety of experimental intricacies, as is the case with other bacterial typing procedures.
Figures
References
-
- Carle G F. Field inversion gel electrophoresis. In: Burmeister M, Ulanowsky L, editors. Methods in molecular biology: pulsed field gel electrophoresis. Totowa, N.J: The Humana Press Inc.; 1992. pp. 3–18. - PubMed
-
- Cookson B D, Aparicio P, Deplano A, Struelens M, Goering R, Marples R. Inter-center comparison of pulsed field gel electrophoresis for the typing of methicillin resistant Staphylococcus aureus. J Med Microbiol. 1996;44:179–184. - PubMed
-
- Hermans P W M, Massadi F, Guebrexhaber H, van Soolingen D, de Haas P E W, Heersma H, de Neeling H, Ayoub A, Portaels F, Frommel D, Zribi M, van Embden J D A. Analysis of the population structure of Mycobacterium tuberculosis in Ethiopia, Tunisia and the Netherlands: usefulness of DNA typing for global tuberculosis epidemiology. J Infect Dis. 1995;171:1504–1513. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

