The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs
- PMID: 9620853
- PMCID: PMC316864
- DOI: 10.1101/gad.12.11.1665
The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs
Abstract
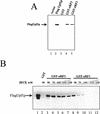
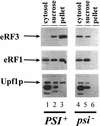
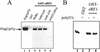
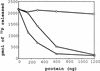
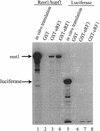
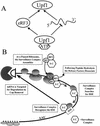
The nonsense-mediated mRNA decay pathway is an example of an evolutionarily conserved surveillance pathway that rids the cell of transcripts that contain nonsense mutations. The product of the UPF1 gene is a necessary component of the putative surveillance complex that recognizes and degrades aberrant mRNAs. Recent results indicate that the Upf1p also enhances translation termination at a nonsense codon. The results presented here demonstrate that the yeast and human forms of the Upf1p interact with both eukaryotic translation termination factors eRF1 and eRF3. Consistent with Upf1p interacting with the eRFs, the Upf1p is found in the prion-like aggregates that contain eRF1 and eRF3 observed in yeast [PSI+] strains. These results suggest that interaction of the Upf1p with the peptidyl release factors may be a key event in the assembly of the putative surveillance complex that enhances translation termination, monitors whether termination has occurred prematurely, and promotes degradation of aberrant transcripts.
Figures





References
-
- Altamura N, Groudinsky O, Dujardin G, Slonimski PP. NAM7 nuclear gene encodes a novel member of a family of helicases with a Z1-6n-ligand motif and is involved in mitochondrial functions in Saccharomyces cerevisiae. J Mol Biol. 1992;224:575–587. - PubMed
-
- Atkin AL, Schenkman LR, Eastham M, Dahlseid JN, Lelivelt MJ, Culbertson MR. Relationship between yeast polyribosomes and Upf proteins required for nonsense mediated mRNA decay. J Biol Chem. 1997;272:22163–22172. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
