Purification and characterization of the nuclear RNase P holoenzyme complex reveals extensive subunit overlap with RNase MRP
- PMID: 9620854
- PMCID: PMC316871
- DOI: 10.1101/gad.12.11.1678
Purification and characterization of the nuclear RNase P holoenzyme complex reveals extensive subunit overlap with RNase MRP
Abstract
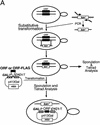
Ribonuclease P (RNase P) is a ribonucleoprotein enzyme that cleaves precursor tRNA transcripts to give mature 5' ends. RNase P in eubacteria has a large, catalytic RNA subunit and a small protein subunit that are required for precursor tRNA cleavage in vivo. Although the eukaryotic holoenzymes have similar, large RNA subunits, previous work in a number of systems has suggested that the eukaryotic enzymes require a greater protein content. We have purified the Saccharomyces cerevisiae nuclear RNase P to apparent homogeneity, allowing the first comprehensive analysis of an unexpectedly complex subunit composition. Peptide sequencing by ion trap mass spectrometry identifies nine proteins that copurify with the nuclear RNase P RNA subunit, totaling 20-fold more protein than in the bacterial enzyme. All of these proteins are encoded by genes essential for RNase P activity and for cell viability. Previous genetic studies suggested that four proteins might be subunits of both RNase P and RNase MRP, the related rRNA processing enzyme. We demonstrate that all four of these proteins, Pop1p, Pop3p, Pop4p, and Rpp1p, are integral subunits of RNase P. In addition, four of the five newly identified protein subunits, Pop5p, Pop6p, Pop7p, and Pop8p, also appear to be shared between RNase P and RNase MRP. Only one polypeptide, Rpr2p, is unique to the RNase P holoenzyme by genetic depletion and immunoprecipitation studies. The large increase in the number of protein subunits over eubacterial RNase P is consistent with an increase in functional complexity in eukaryotes. The degree of structural similarity between nuclear RNase P and RNase MRP suggests that some aspects of their functions in pre-tRNA and pre-rRNA processing pathways might overlap or be coordinated.
Figures


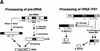
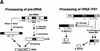


References
-
- Alifano P, Rivellini F, Piscitelli C, Arraiano CM, Bruni CB, Carlomagno MS. Ribonuclease E provides substrates for ribonuclease P-dependent processing of a polycistronic mRNA. Genes & Dev. 1994;8:3021–3031. - PubMed
-
- Altman S, Kirsebom L, Talbot SJ. Recent studies of RNase P. In: Söll D, RajBhandary UL, editors. tRNA: Structure, biosynthesis, and function. Washington, D.C.: American Society of Microbiology; 1995. pp. 67–78.
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Bartkiewicz M, Gold H, Altman S. Identification and characterization of an RNA molecule that copurifies with RNase P activity from HeLa cells. Genes & Dev. 1989;3:488–499. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
