An isoflavonoid-inducible efflux pump in Agrobacterium tumefaciens is involved in competitive colonization of roots
- PMID: 9620959
- PMCID: PMC107810
- DOI: 10.1128/JB.180.12.3107-3113.1998
An isoflavonoid-inducible efflux pump in Agrobacterium tumefaciens is involved in competitive colonization of roots
Abstract
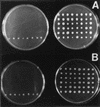
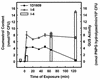
Agrobacterium tumefaciens 1D1609, which was originally isolated from alfalfa (Medicago sativa L.), contains genes that increase competitive root colonization on that plant by reducing the accumulation of alfalfa isoflavonoids in the bacterial cells. Mutant strain I-1 was isolated by its isoflavonoid-inducible neomycin resistance following mutagenesis with the transposable promoter probe Tn5-B30. Nucleotide sequence analysis showed the transposon had inserted in the first open reading frame, ifeA, of a three-gene locus (ifeA, ifeB, and ifeR), which shows high homology to bacterial efflux pump operons. Assays on alfalfa showed that mutant strain I-1 colonized roots normally in single-strain tests but was impaired significantly (P < or = 0.01) in competition against wild-type strain 1D1609. Site-directed mutagenesis experiments, which produced strains I-4 (ifeA::gusA) and I-6 (ifeA::omega-Tc), confirmed the importance of ifeA for competitive root colonization. Exposure to the isoflavonoid coumestrol increased beta-glucuronidase activity in strain I-4 21-fold during the period when coumestrol accumulation in wild-type cells declined. In the same test, coumestrol accumulation in mutant strain I-6 did not decline. Expression of the ifeA-gusA reporter was also induced by the alfalfa root isoflavonoids formononetin and medicarpin but not by two triterpenoids present in alfalfa. These results show that an efflux pump can confer measurable ecological benefits on A. tumefaciens in an environment where the inducing molecules are known to be present.
Figures


Similar articles
-
Characterization of a new Agrobacterium tumefaciens strain from alfalfa.Arch Microbiol. 1998 May;169(5):381-6. doi: 10.1007/s002030050586. Arch Microbiol. 1998. PMID: 9560417
-
Isolation and genetic analysis of an Agrobacterium tumefaciens avirulent mutant with a chromosomal mutation produced by transposon mutagenesis.Biosci Biotechnol Biochem. 1992 Dec;56(12):1924-8. doi: 10.1271/bbb.56.1924. Biosci Biotechnol Biochem. 1992. PMID: 1369092
-
Construction and characterization of Tn5virB, a transposon that generates nonpolar mutations, and its use to define virB8 as an essential virulence gene in Agrobacterium tumefaciens.J Bacteriol. 1993 Feb;175(3):887-91. doi: 10.1128/jb.175.3.887-891.1993. J Bacteriol. 1993. PMID: 8380806 Free PMC article.
-
Isolation and characterization of a new chromosomal virulence gene of Agrobacterium tumefaciens.J Bacteriol. 1993 May;175(10):3208-12. doi: 10.1128/jb.175.10.3208-3212.1993. J Bacteriol. 1993. PMID: 8491736 Free PMC article.
-
Alfalfa (Medicago sativa L.).Methods Mol Biol. 2006;343:301-11. doi: 10.1385/1-59745-130-4:301. Methods Mol Biol. 2006. PMID: 16988354 Review.
Cited by
-
A function of SmeDEF, the major quinolone resistance determinant of Stenotrophomonas maltophilia, is the colonization of plant roots.Appl Environ Microbiol. 2014 Aug;80(15):4559-65. doi: 10.1128/AEM.01058-14. Appl Environ Microbiol. 2014. PMID: 24837376 Free PMC article.
-
Genomic species are ecological species as revealed by comparative genomics in Agrobacterium tumefaciens.Genome Biol Evol. 2011;3:762-81. doi: 10.1093/gbe/evr070. Epub 2011 Jul 27. Genome Biol Evol. 2011. PMID: 21795751 Free PMC article.
-
Antibiotic inducibility of the MexXY multidrug efflux system of Pseudomonas aeruginosa: involvement of the antibiotic-inducible PA5471 gene product.J Bacteriol. 2006 Mar;188(5):1847-55. doi: 10.1128/JB.188.5.1847-1855.2006. J Bacteriol. 2006. PMID: 16484195 Free PMC article.
-
The effect of the Agrobacterium tumefaciens attR mutation on attachment and root colonization differs between legumes and other dicots.Appl Environ Microbiol. 2001 Mar;67(3):1070-5. doi: 10.1128/AEM.67.3.1070-1075.2001. Appl Environ Microbiol. 2001. PMID: 11229893 Free PMC article.
-
Genome sequence of Xanthomonas fuscans subsp. fuscans strain 4834-R reveals that flagellar motility is not a general feature of xanthomonads.BMC Genomics. 2013 Nov 6;14:761. doi: 10.1186/1471-2164-14-761. BMC Genomics. 2013. PMID: 24195767 Free PMC article.
References
-
- Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Barz W. Isolation of rhizosphere bacterium capable of degrading flavonoids. Phytochemistry. 1970;9:1745–1749.
-
- Bolhuis H, van Veen H W, Poolman B, Driessen A J M, Konings W N. Mechanisms of multidrug transporters. FEMS Microbiol Rev. 1997;21:55–84. - PubMed
-
- Cangelosi G A, Best E A, Marinetti G, Nester E W. Genetic analysis of Agrobacterium. Methods Enzymol. 1991;204:384–397. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

