Characterization of the hcnABC gene cluster encoding hydrogen cyanide synthase and anaerobic regulation by ANR in the strictly aerobic biocontrol agent Pseudomonas fluorescens CHA0
- PMID: 9620970
- PMCID: PMC107821
- DOI: 10.1128/JB.180.12.3187-3196.1998
Characterization of the hcnABC gene cluster encoding hydrogen cyanide synthase and anaerobic regulation by ANR in the strictly aerobic biocontrol agent Pseudomonas fluorescens CHA0
Abstract
The secondary metabolite hydrogen cyanide (HCN) is produced by Pseudomonas fluorescens from glycine, essentially under microaerophilic conditions. The genetic basis of HCN synthesis in P. fluorescens CHA0 was investigated. The contiguous structural genes hcnABC encoding HCN synthase were expressed from the T7 promoter in Escherichia coli, resulting in HCN production in this bacterium. Analysis of the nucleotide sequence of the hcnABC genes showed that each HCN synthase subunit was similar to known enzymes involved in hydrogen transfer, i.e., to formate dehydrogenase (for HcnA) or amino acid oxidases (for HcnB and HcnC). These similarities and the presence of flavin adenine dinucleotide- or NAD(P)-binding motifs in HcnB and HcnC suggest that HCN synthase may act as a dehydrogenase in the reaction leading from glycine to HCN and CO2. The hcnA promoter was mapped by primer extension; the -40 sequence (TTGGC ... ATCAA) resembled the consensus FNR (fumarate and nitrate reductase regulator) binding sequence (TTGAT ... ATCAA). The gene encoding the FNR-like protein ANR (anaerobic regulator) was cloned from P. fluorescens CHA0 and sequenced. ANR of strain CHA0 was most similar to ANR of P. aeruginosa and CydR of Azotobacter vinelandii. An anr mutant of P. fluorescens (CHA21) produced little HCN and was unable to express an hcnA-lacZ translational fusion, whereas in wild-type strain CHA0, microaerophilic conditions strongly favored the expression of the hcnA-lacZ fusion. Mutant CHA21 as well as an hcn deletion mutant were impaired in their capacity to suppress black root rot of tobacco, a disease caused by Thielaviopsis basicola, under gnotobiotic conditions. This effect was most pronounced in water-saturated artificial soil, where the anr mutant had lost about 30% of disease suppression ability, compared with wild-type strain CHA0. These results show that the anaerobic regulator ANR is required for cyanide synthesis in the strictly aerobic strain CHA0 and suggest that ANR-mediated cyanogenesis contributes to the suppression of black root rot.
Figures

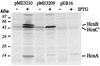
 , genomic DNA from strain CHA0;
, genomic DNA from strain CHA0;  , sites of Tn1725 insertions; ▸, orientation of kanamycin resistance and T7 promoters (PKm and PT7, respectively) on vector plasmids; Tφ, transcription terminator; x, time of Bal 31 digestion (minutes) and extent of deletions created by Bal 31. The HCN phenotype was assessed by qualitative and quantitative tests for HCN production as follows: −, <5 μM HCN; +, wild-type levels of HCN; overproduction of HCN.
, sites of Tn1725 insertions; ▸, orientation of kanamycin resistance and T7 promoters (PKm and PT7, respectively) on vector plasmids; Tφ, transcription terminator; x, time of Bal 31 digestion (minutes) and extent of deletions created by Bal 31. The HCN phenotype was assessed by qualitative and quantitative tests for HCN production as follows: −, <5 μM HCN; +, wild-type levels of HCN; overproduction of HCN.


Similar articles
-
Iron regulation of the hcnABC genes encoding hydrogen cyanide synthase depends on the anaerobic regulator ANR rather than on the global activator GacA in Pseudomonas fluorescens CHA0.Microbiology (Reading). 2000 Oct;146 ( Pt 10):2417-2424. doi: 10.1099/00221287-146-10-2417. Microbiology (Reading). 2000. PMID: 11021918
-
Transcriptional control of the hydrogen cyanide biosynthetic genes hcnABC by the anaerobic regulator ANR and the quorum-sensing regulators LasR and RhlR in Pseudomonas aeruginosa.J Bacteriol. 2000 Dec;182(24):6940-9. doi: 10.1128/JB.182.24.6940-6949.2000. J Bacteriol. 2000. PMID: 11092854 Free PMC article.
-
Mechanism, regulation, and ecological role of bacterial cyanide biosynthesis.Arch Microbiol. 2000 Mar;173(3):170-7. doi: 10.1007/s002039900127. Arch Microbiol. 2000. PMID: 10763748 Review.
-
Persistence and cell culturability of biocontrol strain Pseudomonas fluorescens CHA0 under plough pan conditions in soil and influence of the anaerobic regulator gene anr.Environ Microbiol. 2003 Feb;5(2):103-15. doi: 10.1046/j.1462-2920.2003.00388.x. Environ Microbiol. 2003. PMID: 12558593
-
FNR and its role in oxygen-regulated gene expression in Escherichia coli.FEMS Microbiol Rev. 1990 Aug;6(4):399-428. doi: 10.1111/j.1574-6968.1990.tb04109.x. FEMS Microbiol Rev. 1990. PMID: 2248796 Review.
Cited by
-
Colonization strategies of Pseudomonas fluorescens Pf0-1: activation of soil-specific genes important for diverse and specific environments.BMC Microbiol. 2013 Apr 27;13:92. doi: 10.1186/1471-2180-13-92. BMC Microbiol. 2013. PMID: 23622502 Free PMC article.
-
Stable isotope biomarker breath tests for human metabolic and infectious diseases: a review of recent patent literature.Expert Opin Ther Pat. 2016 Dec;26(12):1393-1398. doi: 10.1080/13543776.2016.1217995. Epub 2016 Aug 10. Expert Opin Ther Pat. 2016. PMID: 27467014 Free PMC article. Review.
-
Draft Genome Sequences of Pseudomonas fluorescens Strains SF39a and SF4c, Potential Plant Growth Promotion and Biocontrol Agents.Genome Announc. 2015 Mar 26;3(2):e00219-15. doi: 10.1128/genomeA.00219-15. Genome Announc. 2015. PMID: 25814613 Free PMC article.
-
Whole Genome, Functional Annotation and Comparative Genomics of Plant Growth-Promoting Bacteria Pseudomonas aeruginosa (NG61) with Potential Application in Agro-Industry.Curr Microbiol. 2022 Apr 23;79(6):169. doi: 10.1007/s00284-022-02845-1. Curr Microbiol. 2022. PMID: 35460384
-
Identification, purification, and characterization of iminodiacetate oxidase from the EDTA-degrading bacterium BNC1.Appl Environ Microbiol. 2001 Feb;67(2):696-701. doi: 10.1128/AEM.67.2.696-701.2001. Appl Environ Microbiol. 2001. PMID: 11157233 Free PMC article.
References
-
- Brunschwig E, Darzins A. A two-component T7 system for the overexpression of genes of Pseudomonas aeruginosa. Gene. 1992;111:35–41. - PubMed
-
- Castric P. Influence of oxygen on the Pseudomonas aeruginosa hydrogen cyanide synthase. Curr Microbiol. 1994;29:19–21.
-
- Castric P A. Hydrogen cyanide, a secondary metabolite of Pseudomonas aeruginosa. Can J Microbiol. 1975;21:613–618. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

