The glucose kinase of Bacillus subtilis
- PMID: 9620975
- PMCID: PMC107826
- DOI: 10.1128/JB.180.12.3222-3226.1998
The glucose kinase of Bacillus subtilis
Abstract
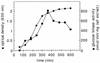
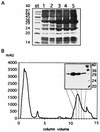
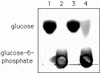
The open reading frame yqgR (now termed glcK), which had been sequenced as part of the genome project, encodes a glucose kinase of Bacillus subtilis. A 1.1-kb DNA fragment containing glcK complemented an Escherichia coli strain deficient in glucose kinase activity. Insertional mutagenesis of glcK resulted in a complete inactivation of glucose kinase activity in crude protein extracts, indicating that B. subtilis contains one major glucose kinase. The glcK gene encodes a 321-residue protein with a molecular mass of 33.5 kDa. The glucose kinase was overexpressed as a fusion protein to a six-His affinity tag and purified to homogeneity. The enzyme had K(m) values for ATP and glucose of 0.77 and 0.24 mM, respectively, and a Vmax of 93 mumol min-1 mg-1. A B. subtilis strain deficient for glucose kinase grew at the same rate on different carbon sources tested, including disaccharides such as maltose, trehalose, and sucrose.
Figures

Similar articles
-
Bacillus subtilis GlcK activity requires cysteines within a motif that discriminates microbial glucokinases into two lineages.BMC Microbiol. 2004 Feb 3;4:6. doi: 10.1186/1471-2180-4-6. BMC Microbiol. 2004. PMID: 15018644 Free PMC article.
-
Molecular characterization of a glucokinase with broad hexose specificity from Bacillus sphaericus strain C3-41.Appl Environ Microbiol. 2007 Jun;73(11):3581-6. doi: 10.1128/AEM.02863-06. Epub 2007 Mar 30. Appl Environ Microbiol. 2007. PMID: 17400775 Free PMC article.
-
Identification and enzymatic characterization of the maltose-inducible alpha-glucosidase MalL (sucrase-isomaltase-maltase) of Bacillus subtilis.J Bacteriol. 1998 May;180(9):2574-8. doi: 10.1128/JB.180.9.2574-2578.1998. J Bacteriol. 1998. PMID: 9573215 Free PMC article.
-
Molecular characterization of glucokinase from Escherichia coli K-12.J Bacteriol. 1997 Feb;179(4):1298-306. doi: 10.1128/jb.179.4.1298-1306.1997. J Bacteriol. 1997. PMID: 9023215 Free PMC article.
-
LOVely enzymes - towards engineering light-controllable biocatalysts.Microb Biotechnol. 2010 Jan;3(1):15-23. doi: 10.1111/j.1751-7915.2009.00140.x. Epub 2009 Aug 24. Microb Biotechnol. 2010. PMID: 21255302 Free PMC article. Review.
Cited by
-
Cloning, expression and characterization of glucokinase gene involved in the glucose-6- phosphate formation in Staphylococcus aureus.Bioinformation. 2013;9(4):169-73. doi: 10.6026/97320630009169. Epub 2013 Feb 21. Bioinformation. 2013. PMID: 23519063 Free PMC article.
-
An update on the review of microbial synthesis of glucosamine and N-acetylglucosamine.World J Microbiol Biotechnol. 2023 Feb 9;39(4):93. doi: 10.1007/s11274-023-03531-5. World J Microbiol Biotechnol. 2023. PMID: 36754899 Review.
-
Systematic genetic dissection of PTS in Vibrio cholerae uncovers a novel glucose transporter and a limited role for PTS during infection of a mammalian host.Mol Microbiol. 2017 May;104(4):568-579. doi: 10.1111/mmi.13646. Epub 2017 Feb 28. Mol Microbiol. 2017. PMID: 28196401 Free PMC article.
-
Thermal stability of glucokinases in Thermoanaerobacter tengcongensis.Biomed Res Int. 2013;2013:646539. doi: 10.1155/2013/646539. Epub 2013 Aug 24. Biomed Res Int. 2013. PMID: 24058911 Free PMC article.
-
The first archaeal ATP-dependent glucokinase, from the hyperthermophilic crenarchaeon Aeropyrum pernix, represents a monomeric, extremely thermophilic ROK glucokinase with broad hexose specificity.J Bacteriol. 2002 Nov;184(21):5955-65. doi: 10.1128/JB.184.21.5955-5965.2002. J Bacteriol. 2002. PMID: 12374829 Free PMC article.
References
-
- Angell S, Schwarz E, Bibb M J. The glucose kinase of Streptomyces coelicolor A3(2): its nucleotide sequence, transcriptional analysis and role in glucose repression. Mol Microbiol. 1992;6:2833–2844. - PubMed
-
- Arora K K, Shenbagamurhi P, Fanciulli M, Pedersen P L. Glucose phosphorylation. J Biol Chem. 1990;265:5324–5328. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

