Influenza virus nucleoprotein interacts with influenza virus polymerase proteins
- PMID: 9621005
- PMCID: PMC110190
- DOI: 10.1128/JVI.72.7.5493-5501.1998
Influenza virus nucleoprotein interacts with influenza virus polymerase proteins
Abstract
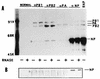
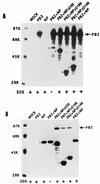
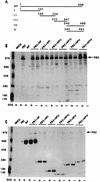
Influenza virus nucleoprotein (NP) is a critical factor in the viral infectious cycle in switching influenza virus RNA synthesis from transcription mode to replication mode. In this study, we investigated the interaction of NP with the viral polymerase protein complex. Using coimmunoprecipitation with monospecific or monoclonal antibodies, we observed that NP interacted with the RNP-free polymerase protein complex in influenza virus-infected cells. In addition, coexpression of the components of the polymerase protein complex (PB1, PB2, or PA) with NP either together or pairwise revealed that NP interacts with PB1 and PB2 but not PA. Interaction of NP with PB1 and PB2 was confirmed by both coimmunoprecipitation and histidine tagging of the NP-PB1 and NP-PB2 complexes. Further, it was observed that NP-PB2 interaction was rather labile and sensitive to dissociation in 0.1% sodium dodecyl sulfate and that the stability of NP-PB2 interaction was regulated by the sequences present at the COOH terminus of NP. Analysis of NP deletion mutants revealed that at least three regions of NP interacted independently with PB2. A detailed analysis of the COOH terminus of NP by mutation of serine-to-alanine (SA) residues either individually or together demonstrated that SA mutations in this region did not affect the binding of NP to PB2. However, some SA mutations at the COOH terminus drastically affected the functional activity of NP in an in vivo transcription-replication assay, whereas others exhibited a temperature-sensitive phenotype and still others had no effect on the transcription and replication of the viral RNA. These results suggest that a direct interaction of NP with polymerase proteins may be involved in regulating the switch of viral RNA synthesis from transcription to replication.
Figures

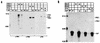



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

