Distinct roles of two binding sites for the bovine papillomavirus (BPV) E2 transactivator on BPV DNA replication
- PMID: 9621032
- PMCID: PMC110248
- DOI: 10.1128/JVI.72.7.5735-5744.1998
Distinct roles of two binding sites for the bovine papillomavirus (BPV) E2 transactivator on BPV DNA replication
Abstract
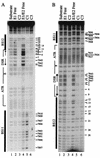
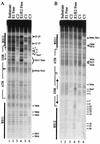
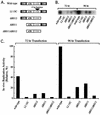
The modulation of DNA replication by transcription factors was examined by using bovine papillomavirus type 1 (BPV). BPV replication in vivo requires two viral proteins: E1, an origin-binding protein, and E2, a transcriptional transactivator. In the origin, E1 interacts with a central region flanked by two binding sites for E2 (BS11 and BS12), of which only BS12 has been reported to be essential for replication in vivo. Using chemical interference and electrophoretic mobility shift assays, we found that the binding of E2 to each site stimulates the formation of distinct E1-origin complexes. A high-mobility C1 complex is formed by using critical E2 contacts to BS12 and E1 contacts to the dyad symmetry element. In contrast, interaction of E2 with the BS11 element on the other origin flank promotes the formation of the lower-mobility C3 complex. C3 is a novel species that resembles C2, a previously identified complex that is replication active and formed by E1 alone. The binding of E1 greatly differs in the C1 and C3 complexes, with E1 in the C1 complex limited to the origin dyad symmetry region and E1 in the C3 complex encompassing the region from the proximal edge of BS11 through the distal edge of BS12. We found that the presence of both E2-binding sites is necessary for wild-type replication activity in vivo, as well as for maximal production of the C3 complex. These results show that in the normal viral context, BS11 and BS12 play separate but synergetic roles in the initiation of viral DNA replication that are dependent on their location within the origin. Our data suggest a model in which the binding of E2 to each site sequentially stimulates the formation of distinct E1-origin complexes, leading to the replication-competent complex.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

