Nonreciprocal packaging of human immunodeficiency virus type 1 and type 2 RNA: a possible role for the p2 domain of Gag in RNA encapsidation
- PMID: 9621049
- PMCID: PMC110391
- DOI: 10.1128/JVI.72.7.5877-5885.1998
Nonreciprocal packaging of human immunodeficiency virus type 1 and type 2 RNA: a possible role for the p2 domain of Gag in RNA encapsidation
Abstract
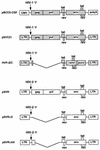
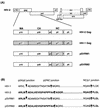
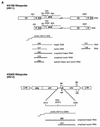
The ability of human immunodeficiency virus types 1 (HIV-1) and 2 (HIV-2) to cross-package each other's RNA was investigated by cotransfecting helper virus constructs with vectors derived from both viruses from which the gag and pol sequences had been removed. HIV-1 was able to package both HIV-1 and HIV-2 vector RNA. The unspliced HIV-1 vector RNA was packaged preferentially over spliced RNA; however, unspliced and spliced HIV-2 vector RNA were packaged in proportion to their cytoplasmic concentrations. The HIV-2 helper virus was unable to package the HIV-1 vector RNA, indicating a nonreciprocal RNA packaging relationship between these two lentiviruses. Chimeric proviruses based on HIV-2 were constructed to identify the regions of the HIV-1 Gag protein conferring RNA-packaging specificity for the HIV-1 packaging signal. Two chimeric viruses were constructed in which domains within the HIV-2 gag gene were replaced by the corresponding domains in HIV-1, and the ability of the chimeric proviruses to encapsidate an HIV-1-based vector was studied. Wild-type HIV-2 was unable to package the HIV-1-based vector; however, replacement of the HIV-2 nucleocapsid by that of HIV-1 generated a virus with normal protein processing which could package the HIV-1-based vector. The chimeric viruses retained the ability to package HIV-2 genomic RNA, providing further evidence for a lack of reciprocity in RNA-packaging ability between the HIV-1 and HIV-2 nucleocapsid proteins. Inclusion of the p2 domain of HIV-1 Gag in the chimera significantly enhanced packaging.
Figures




Similar articles
-
HIV-1 RNA genome packaging: it's G-rated.mBio. 2024 Apr 10;15(4):e0086123. doi: 10.1128/mbio.00861-23. Epub 2024 Feb 27. mBio. 2024. PMID: 38411060 Free PMC article. Review.
-
The nucleocapsid domain is responsible for the ability of spleen necrosis virus (SNV) Gag polyprotein to package both SNV and murine leukemia virus RNA.J Virol. 1999 Nov;73(11):9170-7. doi: 10.1128/JVI.73.11.9170-9177.1999. J Virol. 1999. PMID: 10516024 Free PMC article.
-
The major human immunodeficiency virus type 2 (HIV-2) packaging signal is present on all HIV-2 RNA species: cotranslational RNA encapsidation and limitation of Gag protein confer specificity.J Virol. 2001 Dec;75(24):12058-69. doi: 10.1128/JVI.75.24.12058-12069.2001. J Virol. 2001. PMID: 11711596 Free PMC article.
-
Human immunodeficiency virus types 1 and 2 differ in the predominant mechanism used for selection of genomic RNA for encapsidation.J Virol. 1999 Apr;73(4):3023-31. doi: 10.1128/JVI.73.4.3023-3031.1999. J Virol. 1999. PMID: 10074152 Free PMC article.
-
HIV RNA packaging and lentivirus-based vectors.Adv Pharmacol. 2000;48:1-28. doi: 10.1016/s1054-3589(00)48002-6. Adv Pharmacol. 2000. PMID: 10987087 Review.
Cited by
-
A structural linkage between the dimerization and encapsidation signals in HIV-2 leader RNA.RNA. 2003 Aug;9(8):1007-18. doi: 10.1261/rna.5590603. RNA. 2003. PMID: 12869711 Free PMC article.
-
An extended stem-loop 1 is necessary for human immunodeficiency virus type 2 replication and affects genomic RNA encapsidation.J Virol. 2007 Apr;81(7):3285-92. doi: 10.1128/JVI.02025-06. Epub 2007 Jan 17. J Virol. 2007. PMID: 17229705 Free PMC article.
-
HIV-1 RNA genome packaging: it's G-rated.mBio. 2024 Apr 10;15(4):e0086123. doi: 10.1128/mbio.00861-23. Epub 2024 Feb 27. mBio. 2024. PMID: 38411060 Free PMC article. Review.
-
Encapsidation determinants located downstream of the major splice donor in the maedi-visna virus leader region.J Virol. 2006 Dec;80(23):11743-55. doi: 10.1128/JVI.01284-06. Epub 2006 Sep 13. J Virol. 2006. PMID: 16971429 Free PMC article.
-
Determining the frequency and mechanisms of HIV-1 and HIV-2 RNA copackaging by single-virion analysis.J Virol. 2011 Oct;85(20):10499-508. doi: 10.1128/JVI.05147-11. Epub 2011 Aug 17. J Virol. 2011. PMID: 21849448 Free PMC article.
References
-
- Berkhout B. Structure and function of the human immunodeficiency virus leader RNA. Prog Nucleic Acid Res Mol Biol. 1996;54:1–34. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

