Replication defect of moloney murine leukemia virus with a mutant reverse transcriptase that can incorporate ribonucleotides and deoxyribonucleotides
- PMID: 9621052
- PMCID: PMC110394
- DOI: 10.1128/JVI.72.7.5905-5911.1998
Replication defect of moloney murine leukemia virus with a mutant reverse transcriptase that can incorporate ribonucleotides and deoxyribonucleotides
Abstract
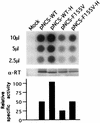
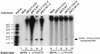
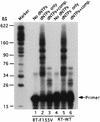
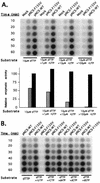
Reverse transcriptase (RT) plays a critical role in retrovirus replication, directing the synthesis of a double- stranded DNA copy of the viral RNA genome. We have previously described a mutant RT of the Moloney murine leukemia virus in which F155 was replaced by valine, and we demonstrated that this substitution allowed the enzyme to incorporate ribonucleotides to form RNA while still retaining its normal ability to incorporate deoxyribonucleotides to form DNA. When introduced into the viral genome, this mutation rendered the virus incapable of replication. Characterization of the mutant virus revealed that the enzyme was still active and able to synthesize minus-strand strong stop DNA and some longer products but failed to make full-length minus-strand DNA. We propose that the failure of the enzyme to complete DNA synthesis in vivo resulted from its ability to incorporate ribonucleotides into the products, which served as inhibitors for DNA synthesis. We also tested seven other amino acid residues for their abilities to substitute for F155 in virus replication; of these, only tyrosine could support virus replication. In an attempt to select for second-site suppressor mutations, the F155V mutant was subjected to random mutagenesis and was used as a parent for the isolation of revertant viruses. Two independent revertants were found to have changed the valine residue at position 155 back to the wild- type phenylalanine. These results suggest that an aromatic ring at this position is important for virus replication.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

