The various Sendai virus C proteins are not functionally equivalent and exert both positive and negative effects on viral RNA accumulation during the course of infection
- PMID: 9621061
- PMCID: PMC110403
- DOI: 10.1128/JVI.72.7.5984-5993.1998
The various Sendai virus C proteins are not functionally equivalent and exert both positive and negative effects on viral RNA accumulation during the course of infection
Abstract
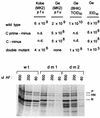
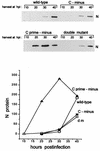
Recombinant Sendai viruses were prepared which cannot express their Cprime, C, or Cprime plus C proteins due to mutation of their respective start codons ([Cprime-minus], [C-minus] and [double mutant], respectively). The [Cprime-minus] and [C-minus] stocks were similar to that of wild-type (wt) virus in virus titer and plaque formation, whereas the double-mutant stock had a much-reduced PFU or 50% egg infective dose/particle ratio and produced very small plaques. Relative to the wt virus infection, the [Cprime-minus] and [C-minus] infections of BHK cells resulted in significantly greater accumulation of viral RNAs, consistent with the known inhibitory effects of the Cprime and C proteins. The double-mutant infection, in contrast, was delayed in its accumulation of viral RNAs; however, once accumulation started, overaccumulation quickly occurred, as in the single-mutant infections. Our results suggest that the Cprime and C proteins both provide a common positive function early in infection, so that only the double mutant undergoes delayed RNA accumulation and exhibits the highly debilitated phenotype. Later in infection, the same proteins appear to act as inhibitors of RNA accumulation. In infections of mice, [Cprime-minus] was found to be as virulent as wt virus whereas [C-minus] was highly attenuated. These results suggest that the Cprime and C proteins cannot be functionally equivalent, since C can replace Cprime for virulence in mice whereas Cprime cannot replace C.
Figures






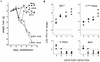
Similar articles
-
Role of primary constitutive phosphorylation of Sendai virus P and V proteins in viral replication and pathogenesis.Virology. 1999 Oct 10;263(1):195-208. doi: 10.1006/viro.1999.9953. Virology. 1999. PMID: 10544094
-
Attenuation of a field Sendai virus isolate through egg-passages is associated with an impediment of viral genome replication in mouse respiratory cells.Arch Virol. 2001;146(5):893-908. doi: 10.1007/s007050170123. Arch Virol. 2001. PMID: 11448028
-
Sendai virus gene start signals are not equivalent in reinitiation capacity: moderation at the fusion protein gene.J Virol. 1999 Nov;73(11):9237-46. doi: 10.1128/JVI.73.11.9237-9246.1999. J Virol. 1999. PMID: 10516032 Free PMC article.
-
The replicative complex of paramyxoviruses: structure and function.Adv Virus Res. 1998;50:101-39. doi: 10.1016/s0065-3527(08)60807-6. Adv Virus Res. 1998. PMID: 9520998 Review. No abstract available.
-
Sendai Virus and a Unified Model of Mononegavirus RNA Synthesis.Viruses. 2021 Dec 9;13(12):2466. doi: 10.3390/v13122466. Viruses. 2021. PMID: 34960735 Free PMC article. Review.
Cited by
-
Sendai virus C proteins counteract the interferon-mediated induction of an antiviral state.J Virol. 1999 Aug;73(8):6559-65. doi: 10.1128/JVI.73.8.6559-6565.1999. J Virol. 1999. PMID: 10400752 Free PMC article.
-
Human parainfluenza virus type 4 is incapable of evading the interferon-induced antiviral effect.J Virol. 2005 Dec;79(23):14756-68. doi: 10.1128/JVI.79.23.14756-14768.2005. J Virol. 2005. PMID: 16282476 Free PMC article.
-
Attenuated and protease-profile modified sendai virus vectors as a new tool for virotherapy of solid tumors.PLoS One. 2014 Mar 5;9(3):e90508. doi: 10.1371/journal.pone.0090508. eCollection 2014. PLoS One. 2014. PMID: 24598703 Free PMC article.
-
Sendai virus C proteins must interact directly with cellular components to interfere with interferon action.J Virol. 2000 Oct;74(19):8823-30. doi: 10.1128/jvi.74.19.8823-8830.2000. J Virol. 2000. PMID: 10982324 Free PMC article.
-
Comparative nucleotide sequence analyses of the entire genomes of B95a cell-isolated and vero cell-isolated measles viruses from the same patient.Virus Genes. 2000;20(3):253-7. doi: 10.1023/a:1008196729676. Virus Genes. 2000. PMID: 10949953
References
-
- Billeter M A, Cattaneo R, Spielhofer P, Kaelin K, Huber M, Schmid A, Baczko K, ter Meulen V. Generation and properties of measles virus mutations typically associated with subacute sclerosing panencephalitis. Ann N Y Acad Sci. 1994;724:367–377. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

