Mouse mammary tumor virus superantigen expression in B cells is regulated by a central enhancer within the pol gene
- PMID: 9621071
- PMCID: PMC110413
- DOI: 10.1128/JVI.72.7.6073-6082.1998
Mouse mammary tumor virus superantigen expression in B cells is regulated by a central enhancer within the pol gene
Abstract
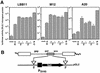
Expression of mouse mammary tumor virus (MMTV)-encoded superantigens in B lymphocytes is required for viral transmission and pathogenesis. The mechanism of superantigen expression from the viral sag gene in B cells is largely unknown, due to problems with detection and quantification of these low-abundance proteins. We have established a sensitive superantigen-luciferase reporter assay to study the expression and regulation of the MMTV sag gene in B-cell lymphomas. The regulatory elements for retroviral gene expression are generally located in the 5' long terminal repeat (LTR) of the provirus. However, we found that neither promoters nor enhancers in the MMTV 5' LTR play a significant role in superantigen expression in these cells. Instead, the essential regulatory regions are located in the pol and env genes of MMTV. We report here that maximal sag expression in B-cell lines depends on an enhancer within the viral pol gene which can be localized to a minimal 183-bp region. Regulation of sag gene expression differs between B-cell lymphomas and pro-B cells, where an enhancer within the viral LTRs is involved. Thus, MMTV sag expression during B-cell development is achieved through the use of two separate enhancer elements.
Figures
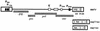





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

