Human herpesvirus 6 open reading frame U12 encodes a functional beta-chemokine receptor
- PMID: 9621074
- PMCID: PMC110416
- DOI: 10.1128/JVI.72.7.6104-6112.1998
Human herpesvirus 6 open reading frame U12 encodes a functional beta-chemokine receptor
Abstract
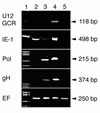
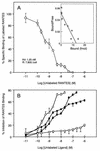
Human herpesvirus 6 (HHV- 6), which belongs to the betaherpesvirus subfamily and infects mainly T cells in vitro, causes acute and latent infections. HHV- 6 contains two genes (U12 and U51) that encode putative homologs of cellular G-protein-coupled receptors (GCR), while three other betaherpesviruses, human cytomegalovirus, murine cytomegalovirus, and human herpesvirus 7, have three, one, and two GCR-homologous genes, respectively. The U12 gene is expressed late in infection from a spliced mRNA. The U12 gene was cloned, and the protein was expressed in cells and analyzed for its biological characteristics. U12 functionally encoded a calcium-mobilizing receptor for beta-chemokines such as regulated upon activation, normal T expressed and secreted (RANTES), macrophage inflammatory proteins 1alpha and 1beta (MIP-1alpha and MIP-1beta) and monocyte chemoattractant protein 1 but not for the alpha-chemokine interleukin-8, suggesting that the chemokine selectivity of the U12 product was distinct from that of the known mammalian chemokine receptors. These findings suggested that the product of U12 may play an important role in the pathogenesis of HHV- 6 through transmembrane signaling by binding with beta-chemokines.
Figures






References
-
- Ablashi D V, Balachandran N, Josephs S F, Hung C L, Krueger G R F, Kramarsky B, Salahuddin S Z, Gallo R C. Genomic polymorphism, growth properties, and immunologic variations in human herpesvirus-6 isolates. Virology. 1991;184:545–552. - PubMed
-
- Ahuja S K, Murphy P M. Molecular piracy of mammarian interleukin-8 receptor type B by herpesvirus saimiri. J Biol Chem. 1993;268:20691–20694. - PubMed
-
- Arvanitakis L, Geras-Raake E, Varma A, Gershengorn M C, Cerarman E. Human herpesvirus KSHV encodes a constitutively active G-protein-coupled receptor linked to cell proliferation. Nature. 1997;358:347–350. - PubMed
-
- Aubin J-T, Agut H, Collandre H, Yamanishi K, Chandran B, Montagnier L, Huraux J-M. Antigenic and genetic differentiation of the two putative types of human herpesvirus 6. J Virol Methods. 1993;41:223–234. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

