Heparan sulfate proteoglycan binding by herpes simplex virus type 1 glycoproteins B and C, which differ in their contributions to virus attachment, penetration, and cell-to-cell spread
- PMID: 9621076
- PMCID: PMC110418
- DOI: 10.1128/JVI.72.7.6119-6130.1998
Heparan sulfate proteoglycan binding by herpes simplex virus type 1 glycoproteins B and C, which differ in their contributions to virus attachment, penetration, and cell-to-cell spread
Abstract
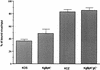
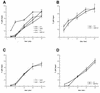
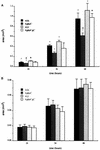
Herpes simplex virus type 1 (HSV-1) mutants defective for envelope glycoprotein C (gC) and gB are highly impaired in the ability to attach to cell surface heparan sulfate (HS) moieties of proteoglycans, the initial virus receptor. Here we report studies aimed at defining the HS binding element of HSV-1 (strain KOS) gB and determining whether this structure is functionally independent of gB's role in extracellular virus penetration or intercellular virus spread. A mutant form of gB deleted for a putative HS binding lysine-rich (pK) sequence (residues 68 to 76) was transiently expressed in Vero cells and shown to be processed normally, leading to exposure on the cell surface. Solubilized gBpK- also had substantially lower affinity for heparin-acrylic beads than did wild-type gB, confirming that the HS binding domain had been inactivated. The gBpK- gene was used to rescue a KOS gB null mutant virus to produce the replication-competent mutant KgBpK-. Compared with wild-type virus, KgBpK- showed reduced binding to mouse L cells (ca. 20%), while a gC null mutant virus in which the gC coding sequence was replaced by the lacZ gene (KCZ) was substantially more impaired (ca. 65%-reduced binding), indicating that the contribution of gC to HS binding was greater than that of gB. The effect of combining both mutations into a single virus (KgBpK-gC-) was additive (ca. 80%-reduced binding to HS) and displayed a binding activity similar to that observed for KOS virus attachment to sog9 cells, a glycosaminoglycan-deficient L-cell line. Cell-adsorbed individual and double HS mutant viruses exhibited a lower rate of virus entry following attachment, suggesting that HS binding plays a role in the process of virus penetration. Moreover, the KgBpK- mutant virus produced small plaques on Vero cells in the presence of neutralizing antibody where plaque formation depended on cell-to-cell virus spread. These studies permitted the following conclusions: (i) the pK sequence is not essential for gB processing or function in virus infection, (ii) the lysine-rich sequence of gB is responsible for HS binding, and (iii) binding to HS is cooperatively linked to the process of efficient virus entry and lateral spread but is not absolutely required for virus infectivity.
Figures

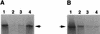



References
-
- Bergström T, Trybala E, Spillmann D. Heparan sulfate and viral tropism. Nat Med. 1997;3:1177. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

