Novel Gag-Pol frameshift site in human immunodeficiency virus type 1 variants resistant to protease inhibitors
- PMID: 9621079
- PMCID: PMC110421
- DOI: 10.1128/JVI.72.7.6146-6150.1998
Novel Gag-Pol frameshift site in human immunodeficiency virus type 1 variants resistant to protease inhibitors
Abstract
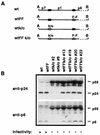
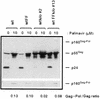
Human immunodeficiency virus type 1 (HIV-1) variants resistant to protease inhibitors have been shown to contain a mutation in the p1/p6 Gag precursor cleavage site. At the messenger RNA level, this mutation generates a U UUU UUU sequence that is reminiscent of the U UUU UUA sequence required for ribosomal frameshifting and Gag-Pol synthesis. To test whether the p1/p6 cleavage site mutation was generating a novel frameshift site, HIV sequences were inserted in translation vectors containing a chloramphenicol acetyltransferase (CAT) reporter gene requiring -1 frameshifting for expression. All sequences containing the original HIV frameshift site supported the synthesis of CAT but expression was increased 3- to 11-fold in the presence of the mutant p1/p6 sequence. When the original frameshift site was abolished by mutation, expression remained unchanged when using constructs containing the mutant p1/p6 sequence, whereas it was decreased 2- to 4.5-fold when using wild-type p1/p6 constructs. Similarly, when introduced into HIV molecular clones, the p1/p6 mutant sequence supported Gag-Pol synthesis and protease activity in the absence of the original frameshift site, indicating that this sequence could also promote ribosomal frameshifting in virus-expressing cells.
Figures

References
-
- Carrillo A, Sham H, Norbeck D, Kempf D, Kohlbrenner W, Plattner J, Leonard J, Molla A. Abstracts of the Fourth Conference on Retroviruses and Opportunistic Infections 1997. Washington. D.C: Infectious Diseases Society of America; 1997. Selection and analysis of HIV-1 variants with increased resistance to ABT-378, a novel protease inhibitor, abstr. 462.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

