The NS5A/NS5 proteins of viruses from three genera of the family flaviviridae are phosphorylated by associated serine/threonine kinases
- PMID: 9621090
- PMCID: PMC110437
- DOI: 10.1128/JVI.72.7.6199-6206.1998
The NS5A/NS5 proteins of viruses from three genera of the family flaviviridae are phosphorylated by associated serine/threonine kinases
Abstract
Phosphorylation of the expressed NS5A protein of hepatitis C virus (HCV), a member of the Hepacivirus genus of the family Flaviviridae, has been demonstrated in mammalian cells and in a cell-free assay by an associated kinase activity. In this report, phosphorylation is also shown for the NS5A and NS5 proteins, respectively, of bovine viral diarrhea virus (BVDV) and yellow fever virus (YF), members of the other two established genera in this family. Phosphorylation of BVDV NS5A and YF NS5 was observed in infected cells, transient expression experiments, and a cell-free assay similar to the one developed for HCV NS5A. Phosphoamino acid analyses indicated that all three proteins were phosphorylated by serine/threonine kinases. Similarities in the properties of BVDV NS5A, YF NS5, and HCV NS5A phosphorylation in vitro further suggested that closely related kinases or the same kinase may phosphorylate these viral proteins. Conservation of this trait among three quite distantly related viruses representing three separate genera suggests that phosphorylation of the NS5A/NS5 proteins or their association with cellular kinases may play an important role in the flavivirus life cycle.
Figures
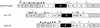


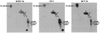



Similar articles
-
Phosphorylation of the hepatitis C virus NS5A protein in vitro and in vivo: properties of the NS5A-associated kinase.J Virol. 1997 Oct;71(10):7187-97. doi: 10.1128/JVI.71.10.7187-7197.1997. J Virol. 1997. PMID: 9311791 Free PMC article.
-
Identification of the major phosphorylation site of the hepatitis C virus H strain NS5A protein as serine 2321.J Biol Chem. 1999 Sep 24;274(39):28011-8. doi: 10.1074/jbc.274.39.28011. J Biol Chem. 1999. PMID: 10488152
-
Hepatitis C virus NS5A protein is phosphorylated by casein kinase II.Biochem Biophys Res Commun. 1999 Apr 21;257(3):777-81. doi: 10.1006/bbrc.1999.0460. Biochem Biophys Res Commun. 1999. PMID: 10208859
-
Phosphorylation of hepatitis C virus NS5A nonstructural protein: a new paradigm for phosphorylation-dependent viral RNA replication?Virology. 2007 Jul 20;364(1):1-9. doi: 10.1016/j.virol.2007.01.042. Epub 2007 Apr 2. Virology. 2007. PMID: 17400273 Review.
-
Non-structural proteins of bovine viral diarrhea virus.Virus Genes. 2022 Dec;58(6):491-500. doi: 10.1007/s11262-022-01914-8. Epub 2022 May 25. Virus Genes. 2022. PMID: 35614328 Free PMC article. Review.
Cited by
-
Atypical Porcine Pestiviruses: Relationships and Conserved Structural Features.Viruses. 2021 Apr 26;13(5):760. doi: 10.3390/v13050760. Viruses. 2021. PMID: 33926056 Free PMC article. Review.
-
CRISPR/Cas9-Mediated Knockout of DNAJC14 Verifies This Chaperone as a Pivotal Host Factor for RNA Replication of Pestiviruses.J Virol. 2019 Feb 19;93(5):e01714-18. doi: 10.1128/JVI.01714-18. Print 2019 Mar 1. J Virol. 2019. PMID: 30518653 Free PMC article.
-
Inhibition of interferon-stimulated JAK-STAT signaling by a tick-borne flavivirus and identification of NS5 as an interferon antagonist.J Virol. 2005 Oct;79(20):12828-39. doi: 10.1128/JVI.79.20.12828-12839.2005. J Virol. 2005. PMID: 16188985 Free PMC article.
-
Interleukin-1 inhibits hepatitis C virus subgenomic RNA replication by activation of extracellular regulated kinase pathway.J Virol. 2003 May;77(9):5493-8. doi: 10.1128/jvi.77.9.5493-5498.2003. J Virol. 2003. PMID: 12692250 Free PMC article.
-
Replication cycle and molecular biology of the West Nile virus.Viruses. 2013 Dec 27;6(1):13-53. doi: 10.3390/v6010013. Viruses. 2013. PMID: 24378320 Free PMC article. Review.
References
-
- Casato M, Agnello V, Pucillo L P, Knight G B, Leoni M, Del Vecchio S, Mazzilli C, Antonelli G, Bonomo L. Predictors of long-term response to high-dose interferon therapy in type II cryoglobulinemia associated with hepatitis C virus infection. Blood. 1997;90:3865–3873. - PubMed
-
- Chambers T J, McCourt D W, Rice C M. Production of yellow fever virus proteins in infected cells: identification of discrete polyprotein species and analysis of cleavage kinetics using region-specific polyclonal antisera. Virology. 1990;177:159–174. - PubMed
-
- Chayama K, Tsubota A, Kobayashi M, Okamoto K, Hashimoto M, Miyano Y, Koike H, Kobayashi M, Koida I, Arase Y, Saitoh S, Suzuki Y, Murashima N, Ikeda K, Kumada H. Pretreatment virus load and multiple amino acid substitutions in the interferon sensitivity-determining region predict the outcome of interferon treatment in patients with chronic genotype 1b hepatitis C virus infection. Hepatology. 1997;25:745–749. - PubMed
-
- Chijiwa T, Mishima A, Hagiwara M, Sano M, Hayashi K, Inoue T, Naito K, Toshioka T, Hidaka H. Inhibition of forskolin-induced neurite outgrowth and protein phosphorylation by a newly synthesized selective inhibitor of cyclic AMP-dependent protein kinase, N-[2-(p-bromocinnamylamino)ethyl]-5-isoquinolinesulfonamide (H-89), of PC12D pheochromocytoma cells. J Biol Chem. 1990;265:5267–5272. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

