Identification of the gene encoding the tryptophan synthase beta-subunit from Chlamydomonas reinhardtii
- PMID: 9625698
- PMCID: PMC34965
- DOI: 10.1104/pp.117.2.455
Identification of the gene encoding the tryptophan synthase beta-subunit from Chlamydomonas reinhardtii
Abstract
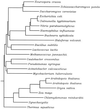
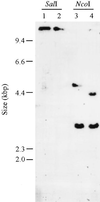
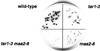
We report the isolation of a Chlamydomonas reinhardtii cDNA that encodes the beta-subunit of tryptophan synthase (TSB). This cDNA was cloned by functional complementation of a trp-operon-deleted strain of Escherichia coli. Hybridization analysis indicated that the gene exists in a single copy. The predicted amino acid sequence showed the greatest identity to TSB polypeptides from other photosynthetic organisms. With the goal of identifying mutations in the gene encoding this enzyme, we isolated 11 recessive and 1 dominant single-gene mutation that conferred resistance to 5-fluoroindole. These mutations fell into three complementation groups, MAA2, MAA7, and TAR1. In vitro assays showed that mutations at each of these loci affected TSB activity. Restriction fragment-length polymorphism analysis suggested that MAA7 encodes TSB. MAA2 and TAR1 may act to regulate the activity of MAA7 or its protein product.
Figures



References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Auerbach S, Gao J, Gussin GN. Nucleotide sequences of the trpI, trpB, and trpA genes of Pseudomonas syringae: positive control unique to fluorescent pseudomonads. Gene. 1993;123:25–32. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

