Stepwise photoinhibition of photosystem II. Studies with Synechocystis species PCC 6803 mutants with a modified D-E loop of the reaction center polypeptide D1
- PMID: 9625701
- PMCID: PMC34968
- DOI: 10.1104/pp.117.2.483
Stepwise photoinhibition of photosystem II. Studies with Synechocystis species PCC 6803 mutants with a modified D-E loop of the reaction center polypeptide D1
Abstract
Several mutant strains of Synechocystis sp. PCC 6803 with large deletions in the D-E loop of the photosystem II (PSII) reaction center polypeptide D1 were subjected to high light to investigate the role of this hydrophilic loop in the photoinhibition cascade of PSII. The tolerance of PSII to photoinhibition in the autotrophic mutant DeltaR225-F239 (PD), when oxygen evolution was monitored with 2,6-dichloro-p-benzoquinone and the equal susceptibility compared with control when monitored with bicarbonate, suggested an inactivation of the QB-binding niche as the first event in the photoinhibition cascade in vivo. This step in PD was largely reversible at low light without the need for protein synthesis. Only the next event, inactivation of QA reduction, was irreversible and gave a signal for D1 polypeptide degradation. The heterotrophic deletion mutants DeltaG240-V249 and DeltaR225-V249 had severely modified QB pockets, yet exhibited high rates of 2,6-dichloro-p-benzoquinone-mediated oxygen evolution and less tolerance to photoinhibition than PD. Moreover, the protein-synthesis-dependent recovery of PSII from photoinhibition was impaired in the DeltaG240-V249 and DeltaR225-V249 mutants because of the effects of the mutations on the expression of the psbA-2 gene. No specific sequences in the D-E loop were found to be essential for high rates of D1 polypeptide degradation.
Figures
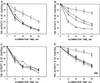
 ) of Synechocystis sp. PCC 6803. Results are the
means of three to five independent experiments from different cell
cultures, and are expressed as a percentage of oxygen-evolution
activity measured before the high-light treatment. Bars denote
) of Synechocystis sp. PCC 6803. Results are the
means of three to five independent experiments from different cell
cultures, and are expressed as a percentage of oxygen-evolution
activity measured before the high-light treatment. Bars denote

 ) strains were illuminated at 1500 μmol
photons m−2 s−1 for 60 min (PI), and the
restoration of DCBQ-dependent oxygen evolution was followed at 40
μmol photons m−2 s−1. All treatments were
performed in the absence of lincomycin.
) strains were illuminated at 1500 μmol
photons m−2 s−1 for 60 min (PI), and the
restoration of DCBQ-dependent oxygen evolution was followed at 40
μmol photons m−2 s−1. All treatments were
performed in the absence of lincomycin.


Similar articles
-
Mutagenesis of the D-E loop of photosystem II reaction centre protein D1. Function and assembly of photosystem II.Plant Mol Biol. 1997 Apr;33(6):1059-71. doi: 10.1023/a:1005765305956. Plant Mol Biol. 1997. PMID: 9154987
-
Comparison of psbO and psbH deletion mutants of Synechocystis PCC 6803 indicates that degradation of D1 protein is regulated by the QB site and dependent on protein synthesis.Biochemistry. 1995 Jul 25;34(29):9625-31. doi: 10.1021/bi00029a040. Biochemistry. 1995. PMID: 7626631
-
Reduced turnover of the D1 polypeptide and photoactivation of electron transfer in novel herbicide resistant mutants of Synechocystis sp. PCC 6803.Eur J Biochem. 1997 Sep 15;248(3):731-40. doi: 10.1111/j.1432-1033.1997.00731.x. Eur J Biochem. 1997. PMID: 9342224
-
Application of low temperatures during photoinhibition allows characterization of individual steps in photodamage and the repair of photosystem II.Photosynth Res. 2007 Nov-Dec;94(2-3):217-24. doi: 10.1007/s11120-007-9184-y. Epub 2007 Jun 7. Photosynth Res. 2007. PMID: 17554634 Review.
-
D1-protein dynamics in photosystem II: the lingering enigma.Photosynth Res. 2008 Oct-Dec;98(1-3):609-20. doi: 10.1007/s11120-008-9342-x. Epub 2008 Aug 16. Photosynth Res. 2008. PMID: 18709440 Review.
Cited by
-
RpaA regulates the accumulation of monomeric photosystem I and PsbA under high light conditions in Synechocystis sp. PCC 6803.PLoS One. 2012;7(9):e45139. doi: 10.1371/journal.pone.0045139. Epub 2012 Sep 14. PLoS One. 2012. PMID: 23024802 Free PMC article.
-
Analysis of the light intensity dependence of the growth of Synechocystis and of the light distribution in a photobioreactor energized by 635 nm light.PeerJ. 2018 Jul 27;6:e5256. doi: 10.7717/peerj.5256. eCollection 2018. PeerJ. 2018. PMID: 30065870 Free PMC article.
-
Multiple light inputs control phototaxis in Synechocystis sp. strain PCC6803.J Bacteriol. 2003 Mar;185(5):1599-607. doi: 10.1128/JB.185.5.1599-1607.2003. J Bacteriol. 2003. PMID: 12591877 Free PMC article.
-
Cyanobacterial psbA gene family: optimization of oxygenic photosynthesis.Cell Mol Life Sci. 2009 Dec;66(23):3697-710. doi: 10.1007/s00018-009-0103-6. Epub 2009 Jul 31. Cell Mol Life Sci. 2009. PMID: 19644734 Free PMC article. Review.
-
Structural insights into photosystem II assembly.Nat Plants. 2021 Apr;7(4):524-538. doi: 10.1038/s41477-021-00895-0. Epub 2021 Apr 12. Nat Plants. 2021. PMID: 33846594 Free PMC article.
References
-
- Aro E-M, Hundal T, Carlberg I, Andersson B. In vitro studies on light-induced inhibition of Photosystem II and D1-protein degradation at low temperatures. Biochim Biophys Acta. 1990;1019:269–275.
-
- Aro E-M, Virgin I, Andersson B. Photoinhibition of photosystem II: inactivation, protein damage and turnover. Biochim Biophys Acta. 1993;1143:113–134. - PubMed
-
- Blubaugh DJ, Cheniae GM. Kinetics of photoinhibition in hydroxylamine-extracted photosystem II membranes: relevance to photoactivation and sites of electron donation. Biochemistry. 1990;29:5109–5118. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

