Interaction of TATA-binding protein with upstream activation factor is required for activated transcription of ribosomal DNA by RNA polymerase I in Saccharomyces cerevisiae in vivo
- PMID: 9632758
- PMCID: PMC108958
- DOI: 10.1128/MCB.18.7.3752
Interaction of TATA-binding protein with upstream activation factor is required for activated transcription of ribosomal DNA by RNA polymerase I in Saccharomyces cerevisiae in vivo
Abstract
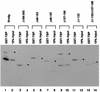
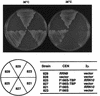
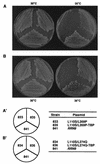
Previous in vitro studies have shown that initiation of transcription of ribosomal DNA (rDNA) in the yeast Saccharomyces cerevisiae involves an interaction of upstream activation factor (UAF) with the upstream element of the promoter, forming a stable UAF-template complex; together with TATA-binding protein (TBP), UAF then recruits an essential factor, core factor (CF), to the promoter, forming a stable preinitiation complex. TBP interacts with both UAF and CF in vitro. In addition, a subunit of UAF, Rrn9p, interacts with TBP in vitro and in the two-hybrid system, suggesting the possible importance of this interaction for UAF function. Using the yeast two-hybrid system, we have identified three mutations in RRN9 that abolish the interaction of Rrn9p with TBP without affecting its interaction with Rrn10p, another subunit of UAF. Yeast cells containing any one of these individual mutations, L110S, L269P, or L274Q, did not show any growth defects. However, cells containing a combination of L110S with one of the other two mutations showed a temperature-sensitive phenotype, and this phenotype was suppressed by fusing the mutant genes to SPT15, which encodes TBP. In addition, another mutation (F186S), which disrupts both Rrn9p-TBP and Rrn9p-Rrn10p interactions in the two-hybrid system, abolished UAF function in vivo, and this mutational defect was suppressed by fusion of the mutant gene to SPT15 combined with overexpression of Rrn10p. These experiments demonstrate that the interaction of UAF with TBP, which is presumably achieved by the interaction of Rrn9p with TBP, is indeed important for high-level transcription of rDNA by RNA polymerase I in vivo.
Figures



References
-
- Bai C, Elledge S J. Gene identification using the yeast two-hybrid system. Methods Enzymol. 1996;273:331–347. - PubMed
-
- Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. - PubMed
-
- Chang J, Kim D-H, Lee S W, Choi K Y, Sung Y C. Transactivation ability of p53 transcriptional activation domain is directly related to the binding affinity to TATA-binding protein. J Biol Chem. 1995;270:25014–25019. - PubMed
-
- Chatterjee S, Struhl K. Connecting a promoter-bound protein to TBP bypasses the need for a transcriptional activation domain. Nature. 1995;374:820–822. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
