TATA binding protein discriminates between different lesions on DNA, resulting in a transcription decrease
- PMID: 9632775
- PMCID: PMC108975
- DOI: 10.1128/MCB.18.7.3907
TATA binding protein discriminates between different lesions on DNA, resulting in a transcription decrease
Abstract
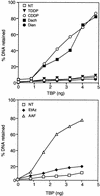
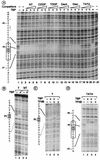
DNA damage recognition by basal transcription factors follows different mechanisms. Using transcription-competition, nitrocellulose filter binding, and DNase I footprinting assays, we show that, although the general transcription factor TFIIH is able to target any kind of lesion which can be repaired by the nucleotide excision repair pathway, TATA binding protein (TBP)-TFIID is more selective in damage recognition. Only genotoxic agents which are able to induce kinked DNA structures similar to the one for the TATA box in its TBP complex are recognized. Indeed, DNase I footprinting patterns reveal that TBP protects equally 4 nucleotides upstream and 6 nucleotides downstream from the A-T (at position -29 of the noncoding strand) of the adenovirus major late promoter and from the G-G of a cisplatin-induced 1,2-d(GpG) cross-link. Together, our results may partially explain differences in transcription inhibition rates following DNA damage.
Figures


Similar articles
-
Transcription factor IIA derepresses TATA-binding protein (TBP)-associated factor inhibition of TBP-DNA binding.J Biol Chem. 1998 Jun 5;273(23):14293-300. doi: 10.1074/jbc.273.23.14293. J Biol Chem. 1998. PMID: 9603936
-
Functional significance of the TATA element major groove in transcription initiation by RNA polymerase II.Nucleic Acids Res. 1997 Nov 1;25(21):4338-45. doi: 10.1093/nar/25.21.4338. Nucleic Acids Res. 1997. PMID: 9336466 Free PMC article.
-
Even-skipped represses transcription by binding TATA binding protein and blocking the TFIID-TATA box interaction.Mol Cell Biol. 1998 Jul;18(7):3771-81. doi: 10.1128/MCB.18.7.3771. Mol Cell Biol. 1998. PMID: 9632760 Free PMC article.
-
Mechanisms of transcriptional activation and repression can both involve TFIID.Philos Trans R Soc Lond B Biol Sci. 1996 Apr 29;351(1339):517-26. doi: 10.1098/rstb.1996.0050. Philos Trans R Soc Lond B Biol Sci. 1996. PMID: 8735274 Review.
-
TBP-associated factors (TAFIIs): multiple, selective transcriptional mediators in common complexes.Trends Biochem Sci. 2000 Feb;25(2):59-63. doi: 10.1016/s0968-0004(99)01527-3. Trends Biochem Sci. 2000. PMID: 10664584 Review.
Cited by
-
The Cellular Response to Transcription-Blocking DNA Damage.Trends Biochem Sci. 2018 May;43(5):327-341. doi: 10.1016/j.tibs.2018.02.010. Trends Biochem Sci. 2018. PMID: 29699641 Free PMC article. Review.
-
On the Role of TATA Boxes and TATA-Binding Protein in Arabidopsis thaliana.Plants (Basel). 2023 Feb 22;12(5):1000. doi: 10.3390/plants12051000. Plants (Basel). 2023. PMID: 36903861 Free PMC article. Review.
-
Structural basis for the sequence-dependent effects of platinum-DNA adducts.Nucleic Acids Res. 2009 May;37(8):2434-48. doi: 10.1093/nar/gkp029. Epub 2009 Mar 2. Nucleic Acids Res. 2009. PMID: 19255091 Free PMC article.
-
Molecular dynamic simulations of cisplatin- and oxaliplatin-d(GG) intrastand cross-links reveal differences in their conformational dynamics.J Mol Biol. 2007 Nov 9;373(5):1123-40. doi: 10.1016/j.jmb.2007.07.079. Epub 2007 Aug 23. J Mol Biol. 2007. PMID: 17900616 Free PMC article.
-
Inhibition of transcription by platinum antitumor compounds.Metallomics. 2009;1(4):280-91. doi: 10.1039/b907567d. Metallomics. 2009. PMID: 20046924 Free PMC article. Review.
References
-
- Alazard R J, Germanier M. Post replication repair in an excision defective strain of Escherichia coli following treatment with cis dichlorodiammineplatinum(II) Biochem Biophys Res Commun. 1982;104:693–700. - PubMed
-
- Bellon S F, Lippard S J. Bending studies of DNA site-specifically modified by cisplatin, trans-diamminedichloroplatinum(II) and cis-[Pt(NH3)2(N3-cytosine)Cl]+ Biophys Chem. 1990;35:179–188. - PubMed
-
- Bohr V A, Smith C A, Okumoto D S, Hanawalt P C. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40:359–369. - PubMed
-
- Boulikas T. DNA lesion-recognizing proteins and the p53 connection. Anticancer Res. 1996;16:225–242. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
