Prohibitin family members interact genetically with mitochondrial inheritance components in Saccharomyces cerevisiae
- PMID: 9632789
- PMCID: PMC108989
- DOI: 10.1128/MCB.18.7.4043
Prohibitin family members interact genetically with mitochondrial inheritance components in Saccharomyces cerevisiae
Abstract
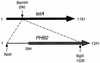
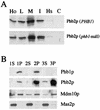
Phb2p, a homolog of the tumor suppressor protein prohibitin, was identified in a genetic screen for suppressors of the loss of Mdm12p, a mitochondrial outer membrane protein required for normal mitochondrial morphology and inheritance in Saccharomyces cerevisiae. Phb2p and its homolog, prohibitin (Phb1p), were localized to the mitochondrial inner membrane and characterized as integral membrane proteins which depend on each other for their stability. In otherwise wild-type genetic backgrounds, null mutations in PHB1 and PHB2 did not confer any obvious phenotypes. However, loss of function of either PHB1 or PHB2 in cells with mitochondrial DNA deleted led to altered mitochondrial morphology, and phb1 or phb2 mutations were synthetically lethal when combined with a mutation in any of three mitochondrial inheritance components of the mitochondrial outer membrane, Mdm12p, Mdm10p, and Mmm1p. These results provide the first evidence of a role for prohibitin in mitochondrial inheritance and in the regulation of mitochondrial morphology.
Figures



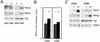

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
