The C-terminal domain of Sin1 interacts with the SWI-SNF complex in yeast
- PMID: 9632800
- PMCID: PMC109000
- DOI: 10.1128/MCB.18.7.4157
The C-terminal domain of Sin1 interacts with the SWI-SNF complex in yeast
Abstract
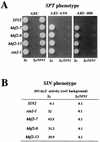
In the yeast Saccharomyces cerevisiae, the SWI-SNF complex has been proposed to antagonize the repressive effects of chromatin by disrupting nucleosomes. The SIN genes were identified as suppressors of defects in the SWI-SNF complex, and the SIN1 gene encodes an HMG1-like protein that has been proposed to be a component of chromatin. Specific mutations (sin mutations) in both histone H3 and H4 genes produce the same phenotypic effects as do mutations in the SIN1 gene. In this study, we demonstrate that Sin1 and the H3 and H4 histones interact genetically and that the C terminus of Sin1 physically associates with components of the SWI-SNF complex. In addition, we demonstrate that this interaction is blocked in the full-length Sin1 protein by the N-terminal half of the protein. Based on these and additional results, we propose that Sin1 acts as a regulatable bridge between the SWI-SNF complex and the nucleosome.
Figures



References
-
- Bernues J, Querol E, Martinez P, Barrios A, Espel E, Llobevas J. Detection by chemical cross-linking of the interaction between high mobility group protein 1 and histone oligomers in free solution. J Biol Chem. 1983;258:11020–11024. - PubMed
-
- Bustin M, Lehn D A, Landsman D. Structural features of the HMG chromosomal proteins and their genes. Biochim Biophys Acta. 1990;1049:231–243. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
