Targeting to transcriptionally active loci by the hydrophilic N-terminal domain of Drosophila DNA topoisomerase I
- PMID: 9632819
- PMCID: PMC109019
- DOI: 10.1128/MCB.18.7.4358
Targeting to transcriptionally active loci by the hydrophilic N-terminal domain of Drosophila DNA topoisomerase I
Abstract
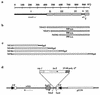
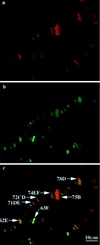
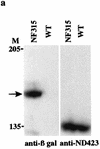
DNA topoisomerase I (topo I) from Drosophila melanogaster contains a nonconserved, hydrophilic N-terminal domain of about 430 residues upstream of the conserved core domains. Deletion of this N terminus did not affect the catalytic activity of topo I, while further removal of sequences into the conserved regions inactivated its enzymatic activity. We have investigated the cellular function of the Drosophila topo I N-terminal domain with top1-lacZ transgenes. There was at least one putative nuclear localization signal within the first 315 residues of the N-terminal domain that allows efficient import of the large chimeric proteins into Drosophila nuclei. The top1-lacZ fusion proteins colocalized with RNA polymerase II (pol II) at developmental puffs on the polytene chromosomes. Either topo I or the top1-lacZ fusion protein was colocalized with RNA pol II in some but not all of the nonpuff, interband loci. However, the fusion proteins as well as RNA pol II were recruited to heat shock puffs during heat treatment, and they returned to the developmental puffs after recovery from heat shock. By immunoprecipitation, we showed that two of the largest subunits of RNA pol II coprecipitated with the N-terminal 315-residue fusion protein by using antibodies against beta-galactosidase. These data suggest that the topo I fusion protein can be localized to the transcriptional complex on chromatin and that the N-terminal 315 residues were sufficient to respond to cellular processes, especially during the reprogramming of gene expression.
Figures







References
-
- Alsner J, Svejstrup J Q, Kjeldsen E, Sorensen B S, Westergaard O. Identification of an N-terminal domain of eukaryotic DNA. J Biol Chem. 1992;267:12408–12411. - PubMed
-
- Ashburner M, Bonner J J. The induction of gene activity in Drosophila by heat shock. Cell. 1979;17:241–254. - PubMed
-
- Bharti A K, Olson M O J, Kufe D M, Rubin E H. Identification of a nucleolin binding site in human topoisomerase I. J Biol Chem. 1996;271:1993–1997. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
