A chloroplast processing enzyme functions as the general stromal processing peptidase
- PMID: 9636172
- PMCID: PMC22651
- DOI: 10.1073/pnas.95.13.7463
A chloroplast processing enzyme functions as the general stromal processing peptidase
Abstract
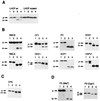
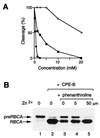
A highly specific stromal processing activity is thought to cleave a large diversity of precursors targeted to the chloroplast, removing an N-terminal transit peptide. The identity of this key component of the import machinery has not been unequivocally established. We have previously characterized a chloroplast processing enzyme (CPE) that cleaves the precursor of the light-harvesting chlorophyll a/b binding protein of photosystem II (LHCPII). Here we report the overexpression of active CPE in Escherichia coli. Examination of the recombinant enzyme in vitro revealed that it cleaves not only preLHCPII, but also the precursors for an array of proteins essential for different reactions and destined for different compartments of the organelle. CPE also processes its own precursor in trans. Neither the recombinant CPE nor the native CPE of chloroplasts process a preLHCPII mutant with an altered cleavage site demonstrating that both forms of the enzyme are sensitive to the same structural modification of the substrate. The transit peptide of the precursor of ferredoxin is released by a single cleavage event and found intact after processing by recombinant CPE and a chloroplast extract as well. These results provide the first direct demonstration that CPE is the general stromal processing peptidase that acts as an endopeptidase. Significantly, recombinant CPE cleaves in the absence of other chloroplast proteins, and this activity depends on metal cations, such as zinc.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

