Myc activates telomerase
- PMID: 9637678
- PMCID: PMC316904
- DOI: 10.1101/gad.12.12.1769
Myc activates telomerase
Abstract
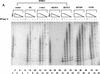
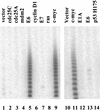
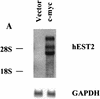
Telomere maintenance has been proposed as an essential prerequisite to human tumor development. The telomerase enzyme is itself a marker for tumor cells, but the genetic alterations that activate the enzyme during neoplastic transformation have remained a mystery. Here, we show that Myc induces telomerase in both normal human mammary epithelial cells (HMECs) and normal human diploid fibroblasts. Myc increases expression of hEST2 (hTRT/TP2), the limiting subunit of telomerase, and both Myc and hEST2 can extend the life span of HMECs. The ability of Myc to activate telomerase may contribute to its ability to promote tumor formation.
Figures










References
-
- Alitalo K, Koskinen P, Makela TP, Saksela K, Sistonen L, Winqvist R. myc oncogenes: Activation and amplification. Biochim Biophys Acta. 1987;907:1–32. - PubMed
-
- Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. - PubMed
-
- Bryan TM, Reddel RR. Telomere dynamics and telomerase activity in in vitro immortalised human cells. Eur J Cancer. 1997;33:767–773. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
