Kappa chain monoallelic demethylation and the establishment of allelic exclusion
- PMID: 9637682
- PMCID: PMC316908
- DOI: 10.1101/gad.12.12.1801
Kappa chain monoallelic demethylation and the establishment of allelic exclusion
Abstract
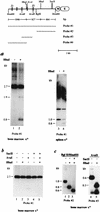
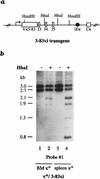
Allelic exclusion in kappa light-chain synthesis is thought to result from a feedback mechanism by which the expression of a functional kappa light chain on the surface of the B cell leads to an intracellular signal that down-regulates the V(D)J recombinase, thus precluding rearrangement of the other allele. Whereas such a feedback mechanism clearly plays a role in the maintenance of allelic exclusion, here we provide evidence suggesting that the initial establishment of allelic exclusion involves differential availability of the two kappa alleles for rearrangement. Analysis of kappa+ B-cell populations and of individual kappa+ B cells that have rearranged only one allele demonstrates that in these cells, critical sites on the rearranged allele are unmethylated, whereas the nonrearranged allele remains methylated. This pattern is apparently generated by demethylation that is initiated at the small pre-B cell stage, on a single allele, in a process that occurs prior to rearrangement and requires the presence in cis of both the intronic and 3' kappa enhancers. Taken together with data demonstrating that undermethylation is required for rearrangement, these results indicate that demethylation may actually underly the process of allelic exclusion by directing the initial choice of a single kappa allele for rearrangement.
Figures




References
-
- Bertram S, Hufert FT, Haefelin DN, von Laer D. Detection of DNA in single cells using an automated cell deposition unit and PCR. BioTechniques. 1995;19:616–620. - PubMed
-
- Betz AG, Milstein C, Gonzalez-Fernandez A, Pannel R, Larson T, Neuberger MS. Elements regulating somatic hypermutation of an immunoglobulin κ gene: Critical role for the intron enhancer/matrix attachment region. Cell. 1994;77:239–248. - PubMed
-
- Chen C, Luning Prak E, Weigert M. Editing disease-associated autoantibodies. Immunity. 1997;6:97–105. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
